All published articles of this journal are available on ScienceDirect.
Multimodal Medical Image Fusion: Techniques, Databases, Evaluation Metrics, and Clinical Applications -A Comprehensive Review
Abstract
Multi-modal Medical Image Fusion (MMIF) is an advancing field at the intersection of medical imaging, data science, and clinical diagnostics. It aims to integrate complementary data from various imaging modalities, such as MRI, CT, and PET, into a single, diagnostically superior composite image. The limitations of unimodal imaging, such as low spatial resolution, insufficient contrast, or incomplete functional characterization, have catalyzed the development of MMIF techniques to enable enhanced visualization, precise diagnosis, and personalized therapeutic planning. This review provides a comprehensive synthesis of the MMIF landscape, categorizing methodologies into five principal domains such as spatial, frequency-based, sparse representation, deep learning, and hybrid approaches. Each technique is critically evaluated for its advantages, limitations, and applicability in clinical settings. Preprocessing, registration, fusion execution, and validation are covered in this review, along with levels of fusion pixel, feature, and decision. The study reviews prominent public databases, including TCIA, OASIS, ADNI, MIDAS, AANLIB, and DDSM, comparing their imaging modalities, disease coverage, file formats, and accessibility. The evaluation of MMIF techniques is systematically addressed, providing a framework for objective performance assessment. An experimental setup is implemented on two datasets to assess the comparative efficacy of selected MMIF techniques utilizing quantitative evaluation variables such as SSIM, entropy, spatial frequency, and mutual information. The results highlight the effectiveness of hybrid and deep learning-based approaches in maintaining both anatomical detail and functional consistency across modalities. The review explores MMIF’s real-world clinical applications, including image-guided neurosurgery, spinal planning, stereotactic radiosurgery, orthopedic implant design, and oncology diagnostics. It also provides insights into future directions, such as explainable AI, federated learning, and integration with robotic surgeries. MMIF offers immense potential yet has limitations like registration errors, computational burdens, generation of artifacts, loss of specific information, and a lack of standardized evaluation metrics. Essentially, the study provides an analytical basis for healthcare experts, scientists, and engineers aiming to develop clinically scalable MMIF systems, which will become indispensable tools for improving diagnostic accuracy, treatment planning, and patient outcomes in modern healthcare.
1. INTRODUCTION
Medical imaging is an essential part of clinical diagnostics because it allows physicians to visualize and evaluate anatomical and physiological structures without surgical intervention. Single imaging approaches are often insufficient to capture all the necessary details, especially when both anatomical and functional aspects need to be evaluated. Magnetic Resonance Imaging (MRI) is known for its high soft tissue resolution but lacks biochemical data detection, whereas Positron Emission Tomography (PET) provides functional metabolic information at the expense of structural precision. Due to these constraints, research has grown significantly in this field. MMIF is defined as the combination of complementary information from various imaging modalities for image enhancement, accuracy, and interpretation. For quality assurance, the fused image should not deviate from the essential characteristic of the input modalities involved and should not be distorted, occluded, or stained with artefacts [1]. For example, combining PET and MR data allows for a detailed depiction of both the anatomy of soft tissues and functional tumor metabolism, which is crucial for oncology and neuroimaging. The use of CT and MRI fusion, as shown in Fig. (1), allows clinicians to have a clear overall overview of the bone structures and soft tissue during treatment planning for radiotherapy and guidance for complex surgical navigations.
The development in the field of medical technology, along with the increasing diversity of medical conditions, substantiates the crucial role of multimodal image fusion. The rise in chronic illnesses makes multimodal imaging a treasured means of understanding the progression of the disease. Despite its merits, there are many challenges for MMIF. The fusion method must accurately preserve the spatial, spectral, and contrast characteristics of the source images. The choice of techniques for co-registration should be proper because images from different modalities, such as PET and MRI, must register precisely, considering the size, resolution, and noise differences [2].
The fusion approaches are categorized into three levels: pixel, feature, and decision, which aim to generate a fused image that enhances visual perception. Traditional medical image fusion techniques are categorized into spatial, transform, and hybrid. The radical change in the automation and performance of methods in MMIF has been fueled by the use of convolutional neural networks, autoencoders, and attention-based architectures [3]. Data-driven learning enables these models to create mature fusion strategies that are more adaptive and generalizable than conventional rule-based models. The application of transformer-based models and GANs has demonstrated positive results for MMIF operations, particularly in PET-MRI fusion and creation of high-resolution medical images [4]. Applications for MMIF include the localization of brain tumors, identifying breast cancer, as well as assessing bone fractures and cardiovascular health. Fused images are also helpful for Computer-Aided Diagnosis (CAD) systems, which, after inputting multimodal data, can generate models that, in turn, increase diagnostic accuracy. Besides, MMIF allows enhanced presentation of key structures and lesions, which are useful in pre-surgical planning, radiotherapy, and interventional procedures [5].
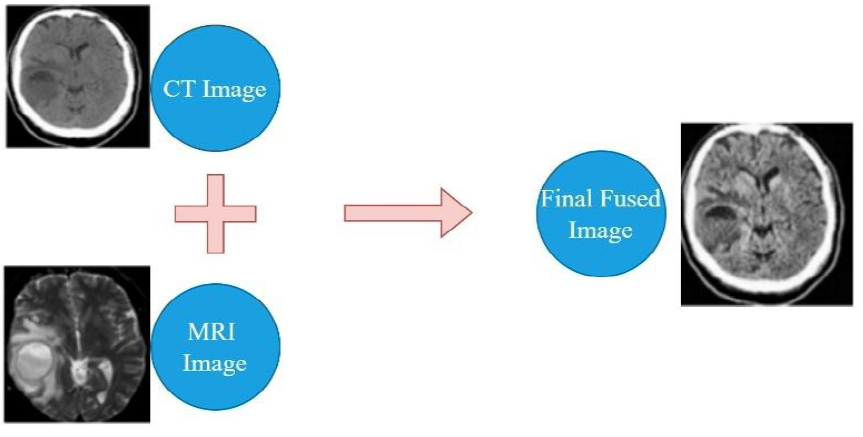
Medical image fusion [2].
This review aims to provide comprehensive information on the multimodal medical image fusion field, encompassing traditional techniques, recent advances, and future trends. The various sections of the review are organized under the following headings.
1. A comparison of major medical imaging modalities.
2. An overview of publicly available multimodal medical image databases, including TCIA, OASIS, ADNI, MIDAS, and AANLIB.
3. MMIF steps: preprocessing, registration, fusion strategy, and performance evaluation.
4. A taxonomy of fusion techniques.
5. Applications of Image Fusion in Clinical Medicine.
6. Image Quality Metrics in Medical Image Fusion.
7. Experimental Set Up and Discussion.
8. Challenges in MMIF deployment.
9. Emerging trends and future directions.
Through this review, we aim to lay a foundational understanding that not only serves current practitioners but also guides future research in designing clinically relevant and scalable MMIF solutions. A comparative analysis of recent review papers on MMIF, emphasizing key criteria such as modality presentation, domain categorization, database accessibility, quantitative evaluation, and clinical applications, is done in Table 1. Figure 2a depicts the growth in publications in this field, and Figure 2b shows the process followed for literature review from 2014 to 2024.
| Review (Year of Publication) | Presentation of Modalities for Imaging | Presented Domains of MMIF |
Publicly Accessible Databases | Quantitative Evaluation Results |
Key Challenges and Future Directions | Clinical Applications |
|---|---|---|---|---|---|---|
| Tirupal et al., 2021 [8] | Yes | Yes | No | Yes | Yes | No |
| Hermessi et al., 2021 [59] | Yes | Yes | No | Yes | Yes | No |
| Haribabu et al. 2022 [5] | Yes | Yes | No | Yes | No | No |
| Saleh et al. 2023 [6] | Yes | Yes | No | Yes | No | No |
| Diwakar et al. 2023 [52] | Yes | Yes | No | Yes | Yes | No |
| Kalamkar and Mary 2023 [2] | Yes | Yes | No | No | Yes | No |
| Khan et al., 2023 [19] | Yes | Yes | Yes | Yes | Yes | No |
| Our Work | Yes | Yes | Yes | Yes | Yes | Yes |
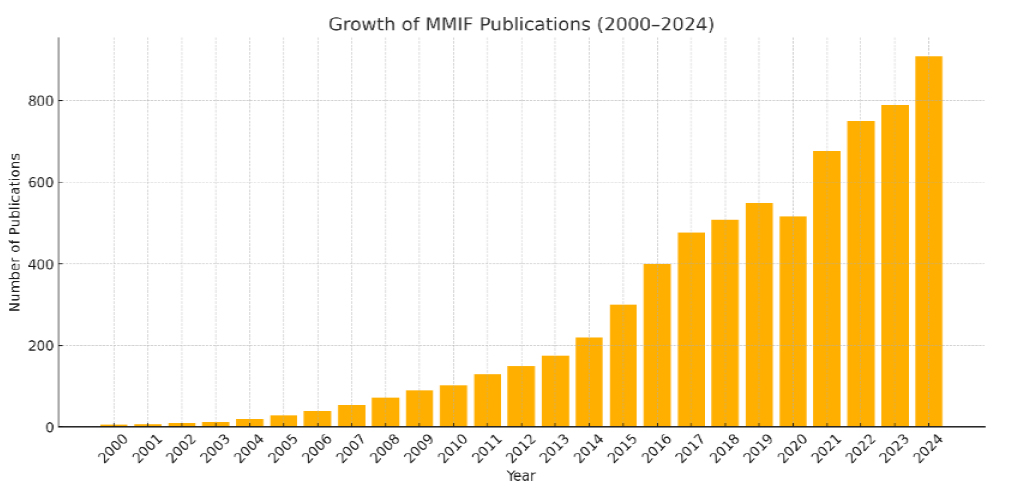
Growth in MMIF publication trend chart (WOS).
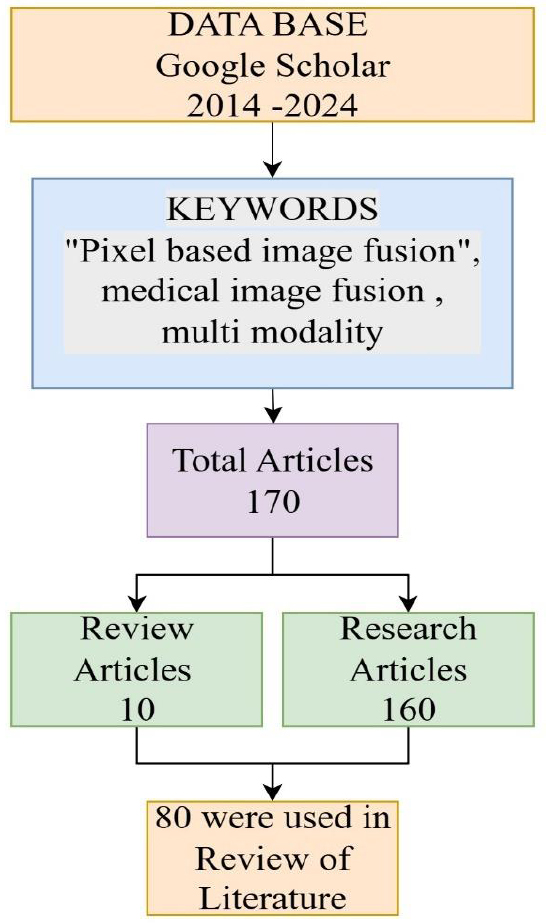
Flowchart illustrating the article selection process for a literature review on pixel-based image fusion in medical imaging, covering publications from 2014 to 2024.
The methodology adopted for this study involved a structured literature search and screening process from 2014–2024. Google Scholar was selected as the principal database due to its comprehensive indexing of scholarly articles across several disciplines, ensuring the inclusion of the most relevant and recent advancements in the field. Specific keywords such as “Pixel-based image fusion,” “medical image fusion,” and “multi-modality” were used. This initial search yielded a total of 170 articles. These were then categorized into review articles (10) and research articles (160) based on their scope and content. The selection process emphasized relevance to the research objectives, specifically focusing on pixel-level fusion methods and their applications in medical imaging. Articles not related to the scope, duplicates, or those with insufficient methodological details were excluded. After this screening, 80 articles were finalized and critically analyzed for the literature review under five different domains, i.e., spatial, transform, deep learning, sparse, and hybrid.
1.1. Imaging Modalities in Medical Image Fusion
Through the fusion of several medical imaging techniques, MMIF manages to gather a variety of information concerning structural and functional features of human bodies. An overview of structural, functional, and multimodal imaging modalities is given in Fig. (3).
X-rays are a fundamental imaging modality and are still highly used for imaging bones, for identifying fractures, infections, tumors, etc., in a two-dimensional image. Computed Tomography (CT) is a method that utilizes ionizing radiation in the form of X-rays for producing cross-sectional three-dimensional images of the body in great detail. It is superior in visualizing bones, revealing internal bleeding, and detecting tumors. CT scans offer rapid and cost-effective imaging, making them a routine choice in trauma evaluations; however, they involve exposure to ionizing radiation [6]. While CT excels in imaging bones compared to MRI, it cannot differentiate soft tissues as effectively.
Magnetic Resonance Imaging (MRI) is a non-invasive technology that uses magnetic fields and radio waves to create high-quality images of soft tissues in the brain, muscles, and other internal organs without the use of ionizing radiation. Owing to its superior image quality and the absence of radiation exposure, MRI is the modality of choice for evaluating brain tumors, spinal cord abnormalities, and ligament injuries [7].
Positron Emission Tomography (PET) is an invasive imaging modality utilizing ionizing radiation to assess metabolic and functional processes within the body. The technique involves administering a radiotracer that emits positrons during radioactive decay. PET is beneficial for oncology, neurology, and cardiology because it detects biochemical changes before anatomical changes are observable. The PET’s weak spatial resolution is complemented by its combination with MRI or CT techniques to enhance anatomical mapping. SPECT and PET use radioactive tracers to measure blood flow and metabolic processes. However, SPECT has less sensitivity and poorer imaging resolution than PET. Despite its limitations, SPECT offers important data for diagnosing cardiac perfusion problems, epilepsy, and bone disorders. When SPECT is combined with CT or MRI, its diagnostic reliability increases significantly, minimizing the effect of its poorer spatial resolution. fMRI uses the measurement of changes in blood oxygenation and flow that correspond to neural activity as an extension of a standard MRI. It is especially evident in brain mapping applications in neuroscience research and surgery planning. Researchers at MMIF often combine fMRI with structural MRI to impart brain function to particular anatomical structures [8]. Table 2 presents the categorization of imaging modalities, such as ultrasound and endoscopy, according to their degree of invasiveness and the extent of patient exposure to ionizing radiation.
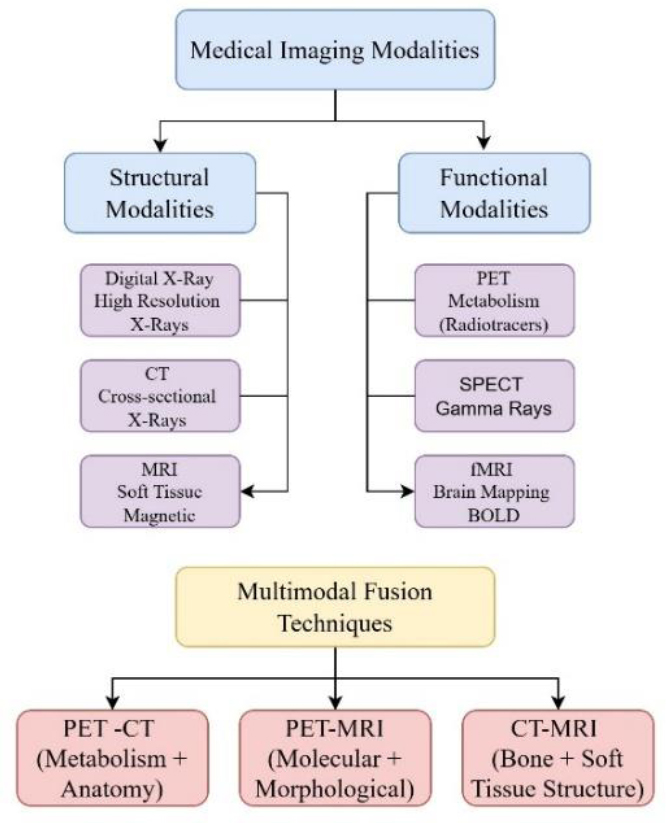
Overview of structural, functional, and multimodal medical imaging modalities.
| Modality | Type of Information | Invasiveness | Ionizing Radiation | Resolution | Typical Use Cases |
|---|---|---|---|---|---|
| MRI | Anatomical (soft tissue) | Non-invasive | No | High spatial | Brain, spinal cord, joints |
| CT | Anatomical (bone/soft tissue) | Non-invasive | Yes | High spatial | Trauma, tumors, lungs |
| PET | Functional (metabolic) | Slightly Invasive (radiotracer) | Yes | Low spatial | Cancer staging, brain disorders |
| SPECT | Functional (blood flow) | Slightly Invasive (radiotracer) | Yes | Low spatial | Cardiology, brain perfusion |
| Ultrasound | Anatomical (real-time soft tissue) | Non-invasive | No | Moderate spatial | Pregnancy, abdomen, heart valves |
| X-ray | Anatomical (bone, dense tissue) | Non-invasive | Yes | Moderate spatial | Fractures, chest infections |
| fMRI | Functional (neural activity) | Non-invasive | No | Moderate spatial | Brain mapping |
| Endoscopy | Visual (surface/internal tissues) | Invasive | No | Very high visual | GI tract visualization, surgical guidance |
2. MULTIMODAL MEDICAL IMAGE DATABASES
The development and validation of Multimodal Medical Image Fusion (MMIF) algorithms require robust, diverse, and accessible imaging datasets. Publicly available datasets offer researchers an opportunity to test their fusion methods across multiple imaging modalities and pathological conditions in a reproducible manner. A variety of high-quality databases have emerged in recent years, as shown in Fig. (4), supporting fusion research across neuroimaging, oncology, cardiology, and more. This section highlights some of the most widely used datasets, TCIA, OASIS, ADNI, MIDAS, AANLIB, and DDSM, and outlines their characteristics, disease focus, modality support, and accessibility.
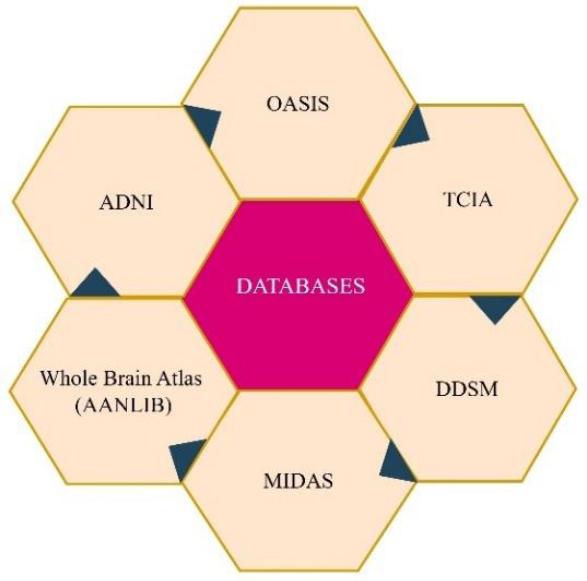
Major publicly available multimodal medical image databases.
2.1. The Cancer Imaging Archive (TCIA)
TCIA appears to be one of the top sources of comprehensive and exhaustively curated cancer imaging data repositories. Under the direction of the National Cancer Institute (NCI), the collection encompasses more than 50,000 richly varied imaging cases CT, MRI, PET, and histopathology. TCIA is a benchmark for assessing fusion algorithms because of its ability to support multimodal imaging research. For example, the Lung-PET-CT-Dx dataset contains synchronized PET and CT scans for the diagnosis of lung cancer, and BraTS contains multimodal MRI (T1, T1-Gd, T2, FLAIR) images annotated for brain tumor research. The platform allows for direct visualization and annotation support through integration with 3D Slicer, ITK-SNAP [9].
2.2. Open Access Series of Imaging Studies (OASIS)
OASIS provides an extensive collection of neuroimaging data, specifically, destined for studies in aging, Alzheimer’s disease, and cognitive deficits. OASIS consists of cross-sectional MRI and PET imaging datasets. OASIS-3, the latest version, contains over 2,000 subjects that had a series of imaging sessions, genetic testing, and clinical testing [10]. The variety of modalities within OASIS datasets makes them ideal for fusion studies. One example is the accuracy of diagnosis of Alzheimer's disease at its earliest stages by using the amyloid PET along with the T1-weighted MRI. Each dataset is represented in the NIfTI format, a reputable neuroimaging standard, and is accompanied by metadata that includes the results of cognitive evaluations.
2.3. Alzheimer’s Disease Neuroimaging Initiative (ADNI)
ADNI is a collaborative investigation for the study of the progress of Alzheimer’s disease through imaging and clinical data. It provides rich longitudinal data drawn from several imaging and biomarker modalities such as MRI, FDG-PET, amyloid PET, and CSF biomarkers. The initiative tracks more than 1,700 participants through various stages of cognitive decline. ADNI research subjects are characterized at three cognitive levels such as cognitively normal, MCI, and Alzheimer’s disease. The large temporal resolution of ADNI allows the study of temporal dynamics of brain change represented by other imaging technologies. In many ADNI data-based investigations, researchers use fusion methods to predict progression from MCI to Alzheimer’s by combining structural MRI with metabolic PET information [11]. Researchers can obtain data from the ADNI through an application process, in the form of DICOM and NIfTI, via the Laboratory of Neuro Imaging (LONI) platform.
2.4. Medical Image Data Archive System (MIDAS)
MIDAS, an adaptable data management system, was developed by Kitware, which serves the area of imaging datasets, including the realms of radiology, pathology, and ultrasound. The system’s flexible architecture enables the integration and storage of 2D and 3D imaging data together with corresponding clinical and demographic records. The uniqueness of MIDAS is its capability to integrate custom plugins and tools, which is useful for researchers who work with end-to-end pipelines for image fusion and segmentation. Some of the commonly used datasets in MMIF research include Head-Neck Cancer CT-MRI and Cardiac MR + Ultrasound [12]. MIDAS can handle DICOM, RAW, and MetaImage (.mha).
2.5. Digital Database for Screening Mammography (DDSM)
The DDSM repository exists with the objective of large-scale breast detection, predominantly facilitated by X-ray mammograms. Although the repository originally concentrated on one modality, the recent additions, such as INbreast and CBIS-DDSM, have included histopathology and ultrasound data for supporting multimodal analysis. The DDSM collection contains more than 2,500 studies, several of which are annotated with information about mass boundaries. The DDSM’s usefulness for MMIF lies in its synthesis of mammographic features with pathological confirmation, supporting the integration of image and diagnostic data [13].
2.6. Annotated Alzheimer Neuroimaging Library (AANLIB)
AANLIB was created as a dataset specific to assisting multimodal fusion and classification research for Alzheimer’s disease. It includes thousands of cases with T1-MRI, FLAIR, PET, and neurocognitive records. Unlike large datasets such as OASIS or ADNI, AANLIB provides images that are prepared for fusion (preprocessing corrects for skull stripping and registration). These standardized images significantly streamline the work in the MMIF workflows. The AANLIB’s corresponding images are archived in NIfTI format, and its accompanying ground truth labels are provided for employing it in supervised learning procedures. The open access for academic users makes it suitable for evaluating deep learning fusion algorithms [14]. The source images of the brain from AANLIB in axial, sagittal, and coronal sections are presented in Fig. (5).
2.7. Dataset Summary and Usage Patterns
The most widely used fusion modalities across these datasets include MRI+PET, MRI+CT, and PET+CT, reflecting the clinical demand for combining anatomical and functional insights. For example, ADNI and OASIS predominantly use MRI+PET, while TCIA and MIDAS offer CT+PET or MRI+CT combinations. Table 3 provides an overview of the available databases with respect to imaging modalities, anatomical regions, and file formats. Regarding disease focus, the datasets are primarily oriented toward the following pathological conditions:
- Neurodegenerative diseases: OASIS, ADNI, AANLIB
- Cancer imaging: TCIA, DDSM, MIDAS
- Cardiac and head-neck imaging: MIDAS
- Neurofunctional tasks: AANLIB, OASIS
3. MMIF PROCESS FLOW AND FUSION LEVELS
Multimodal Medical Image Fusion (MMIF) is a complex, multistage process designed to integrate complementary information from multiple imaging modalities into a single, more informative representation. This process is typically divided into four primary stages such as preprocessing, registration, fusion, and validation. Each stage involves distinct technical considerations and affects the overall quality and clinical reliability of the fused output. Furthermore, fusion can be performed at different abstraction levels, pixel, feature, and decision, each offering specific benefits and trade-offs. Understanding this full workflow is critical for designing and evaluating robust MMIF systems.
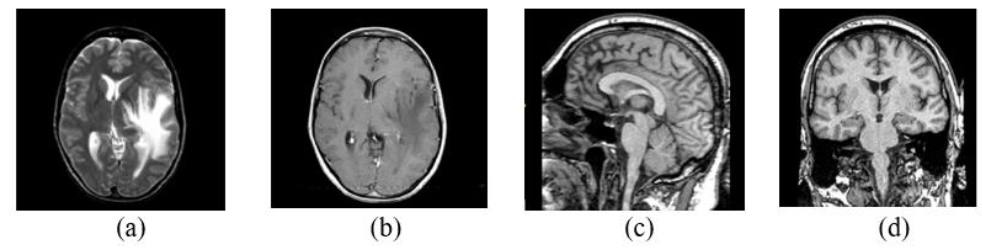
AANLIB Source images of the brain in axial (a) and (b), sagittal (c), and coronal sections (d) [14].
| Database | Modalities | Target Organs / Systems | File Formats | Access Type |
|---|---|---|---|---|
| TCIA | CT, MRI, PET, Histopathology | Brain, Lung, Breast, Prostate | DICOM, NIfTI | Open (via API & GUI) |
| ADNI | MRI, PET, CSF | Brain (Neurodegeneration) | DICOM, NIfTI | Request-based (LONI access) |
| OASIS | MRI, PET | Brain (Aging, Dementia) | NIfTI | Open (with Data Use Agreement) |
| AANLIB | MRI, PET, FLAIR | Brain (Alzheimer’s) | NIfTI | Open (Academic Use) |
| MIDAS | CT, MRI, Ultrasound | Head & Neck, Heart, Abdomen | DICOM, MHA, RAW | Open (Kitware tools) |
| DDSM | Mammography, Histopathology | Breast | LJPEG, DICOM | Open (Preprocessing scripts available) |
Steps in the process of image fusion [15].
3.1. Preprocessing
Preprocessing serves as the foundation of MMIF and involves preparing images for further stages by enhancing quality and ensuring uniformity across modalities. This stage includes noise reduction, intensity normalization, contrast enhancement, resolution adjustment, and format conversion. Since different imaging modalities have different spatial resolutions, acquisition angles, and noise characteristics, preprocessing plays a vital role in aligning these disparities. For instance, Magnetic Resonance Imaging (MRI) often exhibits inhomogeneous intensity, while Computed Tomography (CT) is prone to beam-hardening artifacts. Normalizing these differences using histogram equalization or z-score normalization improves the effectiveness of subsequent fusion steps. Additionally, many public datasets like AANLIB provide preprocessed images, including skull stripping and spatial standardization, reducing the preprocessing burden on researchers [15]. Advanced preprocessing techniques such as total variation filtering, anisotropic diffusion, and rolling guidance filters have been adopted in recent MMIF research to preserve structural details while eliminating noise. These techniques are especially beneficial in fusion scenarios involving low-quality or low-contrast images. The process of image fusion is shown in Fig. (6).
3.2. Image Registration
Registration is the process of spatially aligning two or more images of the same anatomical region, but from different modalities. It compensates for differences in image scale, orientation, and position, ensuring that corresponding anatomical structures overlap accurately. The accuracy of registration directly affects the fidelity of the final fused image, especially in pixel- and feature-level fusion.
Image registration methods are typically categorized into rigid, affine, and non-rigid (deformable) techniques. Rigid registration handles only translation and rotation, while affine registration includes scaling and shearing. Non-rigid registration addresses complex deformations and is often necessary when fusing modalities like PET and MRI due to organ motion or different acquisition geometries. Techniques such as mutual information maximization, normalized cross-correlation, and landmark-based mapping are commonly employed. Deep learning-based registration models, especially U-Net architectures trained on spatial transformer networks, are gaining popularity due to their speed and robustness. These methods can perform unsupervised, real-time registration even in challenging clinical settings.
3.3. Image Fusion Techniques
The core of MMIF is the fusion process itself, which combines information from registered images into a single image. Fusion techniques are classified based on the level at which integration occurs, as described in Fig. (7).
3.3.1. Pixel-level Fusion
Pixel-level fusion is the most straightforward approach, where corresponding pixels from the source images are directly combined using arithmetic or logical operations. Common methods include weighted averaging, maximum selection, and wavelet-based combination. These methods are computationally efficient but highly sensitive to registration errors and noise. Recent enhancements at this level involve multiscale transforms such as NSCT (Non-subsampled Contourlet Transform), DWT, and shearlet transforms, which improve spatial-frequency localization and reduce artifacts [16].
3.3.2. Feature-level Fusion
Feature-level fusion focuses on processing and integrating visual features that are more relevant than individual pixels. This strategy involves independently extracting high-level characteristics such as edges, textures, and shapes from the source images and then using a fusion method on them. This level offers better robustness against misregistration and provides more semantically meaningful outputs, leading to enhanced visual perception, decision-making precision. Techniques like SIFT (Scale-Invariant Feature Transform), Gabor filters, and gradient domain methods are widely used for this purpose.
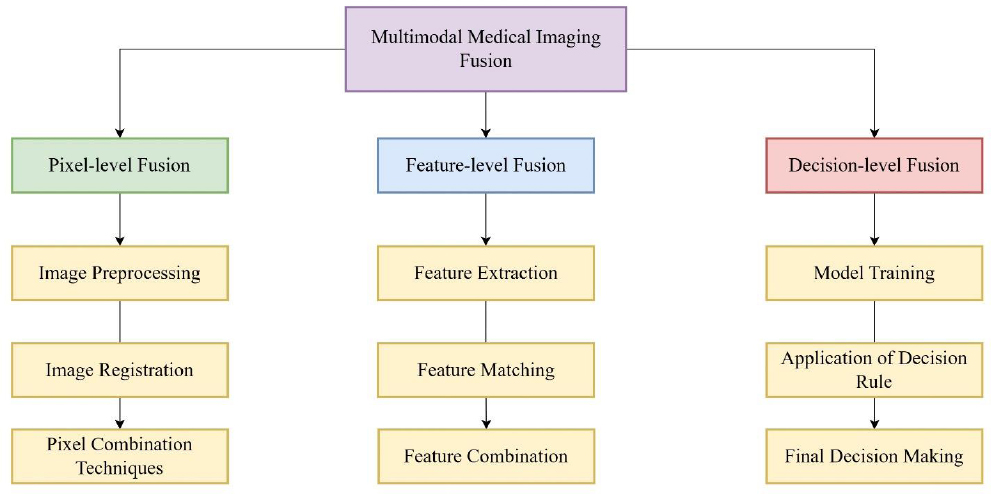
Levels of multimodal medical image fusion.
| Fusion Level | Description | Techniques | Advantages | Limitations | Typical Applications |
|---|---|---|---|---|---|
| Pixel-Level | Combines corresponding pixels from source images using arithmetic or transform-based methods | Averaging, Maximum Selection, DWT, NSCT, Shearlet | Preserves fine structural details; computationally simple | Highly sensitive to noise and registration errors | MRI-CT fusion in brain imaging; PET-CT for tumor mapping |
| Feature-Level | Extracts and fuses intermediate features such as edges, texture, and gradients before reconstruction | SIFT, CNN features, sparse representation, PCA | Robust to alignment errors; preserves semantic content | High computational complexity; depends on effective feature extraction | Alzheimer’s analysis using MRI-PET; tumor segmentation from hybrid MRI |
| Decision-Level | Fusion occurs after independent modality analysis at the classification or prediction stage. | Majority voting, Bayesian inference, ensemble fusion | Allows modality-specific models; lower dependence on pixel accuracy | Does not generate a fused image; interpretability is reduced | CAD systems for tumor detection, AI-based diagnosis integration |
Feature-level fusion can be categorized according to the nature of the methodologies used and the combined features. These techniques can be broadly categorized into sparse representation methods and clustering-based methods. Sparse techniques express images as sparse vectors in a dictionary. These methods typically divide source images into patches, organize them into vectors, and then perform the fusion process. Clustering algorithms, including Quantum Particle Swarm Optimization and Fuzzy C-means, can split feature spaces and provide weighting factors for fusion.
3.3.3. Decision-level Fusion
At the decision level, fusion occurs after separate processing and classification of input images. This is commonly used in Computer-Aided Diagnosis (CAD) systems and is especially relevant in AI-based clinical workflows. Methods for decision-level fusion include majority voting, Bayesian inference, Dempster-Shafer theory, and ensemble learning techniques, which weigh individual decisions to derive a robust outcome. While decision-level fusion is less sensitive to registration errors and allows for modality-specific preprocessing and modeling, it lacks spatial resolution in the final output. It is generally unsuitable when a fused image is required for direct interpretation [17]. A brief comparison of these three, along with the scope of application, is summarized in Table 4.
3.4. Validation and Evaluation
Validation assesses the quality and clinical utility of the fused image using objective metrics and, where possible, expert evaluation. The most commonly used metrics include:
- Structural Similarity Index (SSIM): Measures perceived image quality and structural preservation.
- Peak Signal-to-Noise Ratio (PSNR): Evaluates image fidelity based on pixel intensity.
- Mutual Information (MI): Quantifies the amount of shared information between fused and source images.
- Entropy (EN): Reflects information richness in the fused image.
- Edge Preservation Index (EPI) and Spatial Frequency (SF): Measure edge clarity and texture detail [18].
Visual comparison remains essential in clinical validation. Radiologists or clinical experts often review Fusion outputs to assess diagnostic relevance and interpretability. Increasingly, evaluation also includes testing downstream tasks such as segmentation, classification, and localization to assess the practical benefits of fusion. MMIF is a multi-stage process where each phase requires tailored algorithms and quality checks to ensure robust performance, from preprocessing to validation. Fusion can be applied at different levels depending on the application, with each level offering unique advantages. Recent advances, especially in deep learning and transformer models, have improved registration accuracy and fusion robustness, allowing real-time, scalable implementations in clinical environments. The modular nature of the MMIF makes it adaptable, allowing hybrid strategies that combine pixel, feature, and decision-level fusion to maximize clinical efficacy [19]. The various steps in the entire workflow are depicted in Fig. (8).
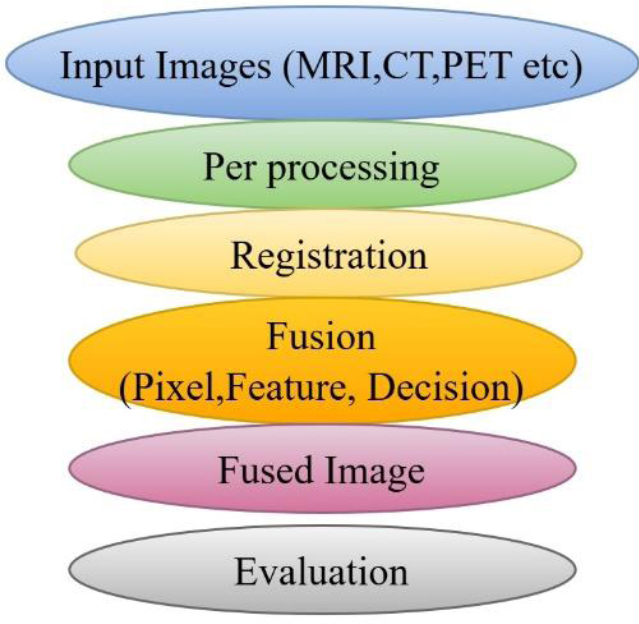
MMIF workflow [19].
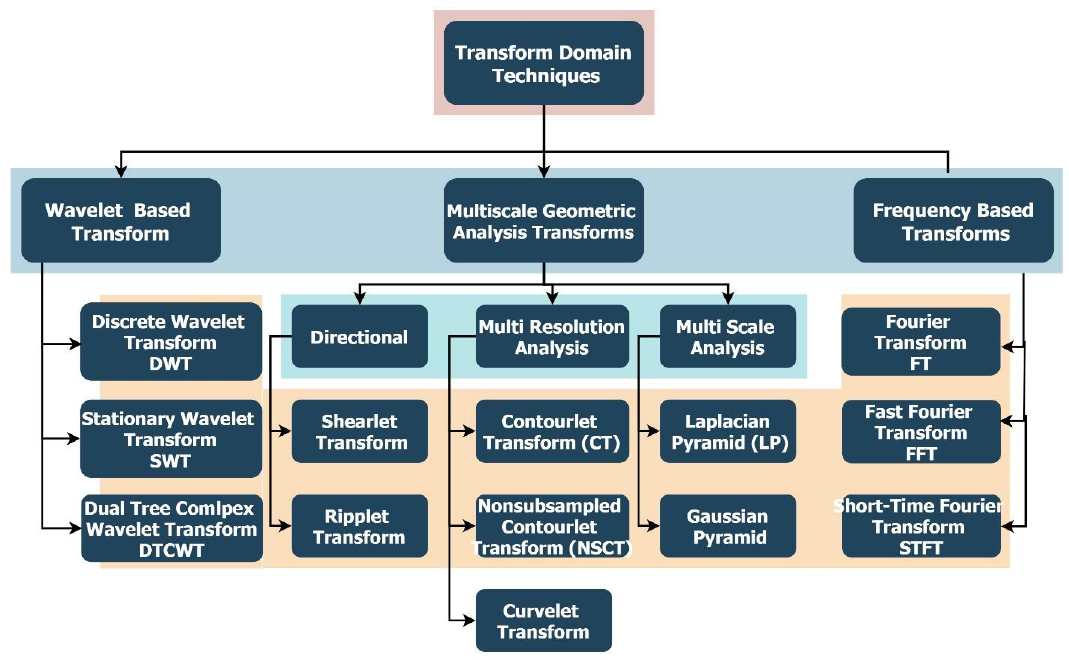
4. FUSION DOMAINS AND TECHNIQUES
These MMIF methods are used in diverse computational realms, which impart unique advantages. The choice of domain affects the efficiency, quality, and real-world applicability of fusion results. The prominent domains and their respective techniques are described in Fig. (9).
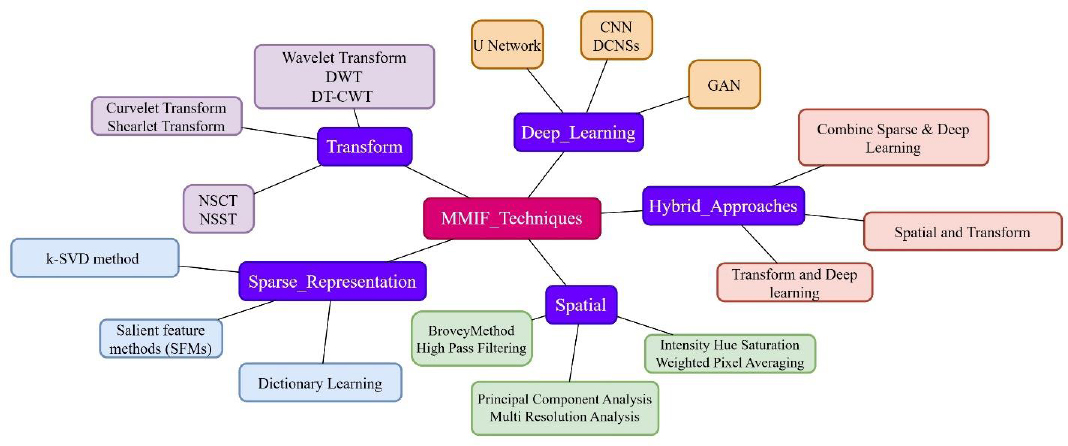
Various domains of multimodal medical image fusion along with techniques.
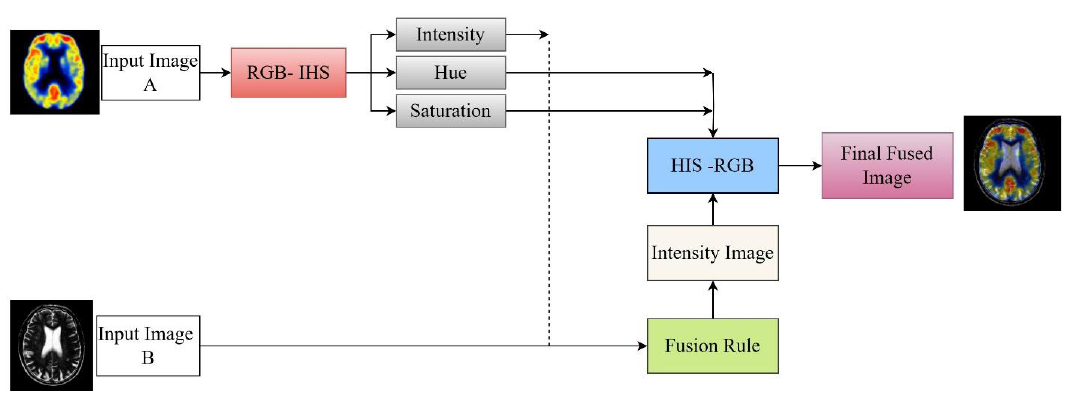
Fusion of SPECT and MRI images using IHS in the spatial domain [19].
This section presents a structured review of the dominant fusion domains under five headings:
- Spatial Domain
- Frequency /Transform Domain
- Sparse representation
- Deep learning, and
- Hybrid Domain.
4.1. Spatial Domain
Image fusion in the spatial domain is distinguished by simple computation and sound handling, which can be easily used and processed. In this process, image fusion is performed using methods that manipulate pixel values without first transforming them into frequency domain formats. Conventional spatial fusion methods include Principal Component Analysis (PCA), Intensity-Hue-Saturation (IHS) mapping, Brovey transformation, and high-pass filtering. Such methods have the advantage of simplicity, faster execution times, and more vivid color expression, making them suitable for real-time cases and applications where computational power is limited. For instance, SPECT-MRI fusion has opportune over the IHS-based methods, which enhance the blending of anatomical and functional views without involving complex computational routines, as shown in Fig. (10).
Spatial domain methods tend to cause edge softening, spectral distortion, and weak noise tolerance. Since these techniques lack frequency composition, they cannot capture subtle textural and high-frequency details, and thus their role in complex applications such as brain tumor localization or microvascular imaging cannot be fully realized. Initial Studies by Baraiya and Gagnani [20] and Parekh et al. [21] highlighted traditional techniques like Principal Component Analysis (PCA) and Brovey Transform, which are utilized in fields such as remote sensing and preliminary diagnostic systems. These methods highlighted fundamental spatial integration while exposing significant problems, including spatial distortions and inadequate spectral preservation. Morris and Rajesh [22] recognized the constraints of static fusion rules, advocating for image-adaptive approaches. Bhuvaneswari and Dhanasekaran [23] identified that conventional spatial domain techniques diminish image contrast, prompting the advancement of transform-domain solutions.
Li et al. [24] and Du et al. [25] made significant advancements by incorporating multi-scale transforms and edge-preserving filtering, thereby enhancing both structural integrity and contrast preservation. Their efforts established the foundation for hybrid spatial-transform techniques. Zhan et al. [26] tackled brain image fusion by implementing guided filtering and spatial gradient-based enhancements, which demonstrated enhanced performance in high-detail areas like cerebral tissue and lesions. Kotian et al. [27] conducted a comparative analysis, suggesting that a spatial/wavelet hybrid approach, which combined both spatial and spectral attributes, is a recommended method for general Multi-Image (MI) fusion. From 2018–2023, the field underwent substantial maturation when researchers such as Liu et al. [28], Na et al. [29], Saboori and Birjandtalab [30], and Pei et al. [31] employed wavelet transforms, multi-resolution decomposition, and guided filtering to preserve feature layers and minimize distortion. They attained significant success in applications including PET-MRI, CT-MRI fusion, and brain pathology analysis. Tan et al. [32], Chen et al. [33], and Deepali Sale [34] focused on enhancing low-complexity algorithms for rapid implementation in real-time systems, whereas Kong et al. [35], Li et al. [36], Feng et al. [37], and Zhang et al. [38] introduced sophisticated spatial techniques such as Framelet Transform, quasi-bilateral filtering, and structural dissimilarity metrics. These recent methodologies show enhanced performance in terms of structural retention, contrast preservation, and computational efficiency relative to State-of-the-Art (SOTA) fusion techniques. The progression of spatial domain techniques indicated a trend towards hybridization, multi-scale modeling, and adaptive spatial decision-making, thereby enhancing robustness in clinical applications such as lesion localization, radiotherapy planning, and neuroimaging diagnostics. Table 5 depicts the fusion strategy and area of application in the spatial domain.
4.2. Transform Domain
The transform or frequency domain has emerged as a powerful approach to mitigate several challenges associated with secure image fusion. Leading up to these transformations is the conversion of images using conversion algorithms such as the Fourier Transform, Discrete Wavelet Transform (DWT), Non-Subsampled Contourlet Transform (NSCT), or Laplacian Pyramid method. These changes separate images into separate spatial-frequency components, thus allowing for a more accurate separation of structural vs. textural aspects. Thereafter, when fusion is carried out in the transform domain, the reconstructed final image features an inverse transformation. Since frequency-based approaches excel at maintaining edge sharpness and texture information, they are suitable for clinical endeavors involving precise structure separation, such as identifying tumor edges. The basic process of decomposition and application of the inverse transform is shown in Fig. (11).
| Author (Year) | Technique | Method | Fusion Strategy | Area of Application |
|---|---|---|---|---|
| Baraiya & Gagnani (2014) [20] | PCA, direct pixel integration | Classical Spatial | Basic fusion using PCA for improved interpretability | Computer vision, medical imaging |
| Parekh et al. (2014) [21] | Brovey Transform, Color Model | Spatial | Color normalization and red channel enhancement; prone to spatial distortion | Remote sensing, radiology |
| Morris & Rajesh (2015) [22] | Pixel arithmetic (avg, add, subtract) | Spatial | Emphasized input-based method selection; basic fusion is not always optimal | Diagnostic imaging |
| Bhuvaneswari & Dhanasekaran (2016) [23] | Spatial vs. Transform | Comparative Study | Highlighted contrast loss in spatial fusion; promoted hybrid alternatives | MRI-CT, PET fusion |
| Li et al. (2017) [24] | Multi-scale + edge preserving filter | Spatial-Transform Hybrid | Proposed framework to preserve detail in structural fusion | Clinical diagnostics |
| Du et al. (2017) [25] | Local Laplacian filtering + multi-scale framework | Spatial | Used predefined features for distortion-free PET-SPECT fusion | Brain imaging |
| Zhan et al. (2017) [26] | Fast gradient filtering + morphological closure | Gradient-Based Spatial | Introduced fast structure-preserving filters; reduced execution time | Multi-organ fusion |
| Kotian et al. (2017) [27] | Comparative analysis | Spatial / Wavelet Hybrid | Recommended combination of spatial and spectral attributes | General MI fusion |
| Liu et al. (2018) [28] | Multi-scale joint decomposition + shearing filters | Transform Hybrid | Directional coefficients for high-detail retention | Brain functional imaging |
| Na et al. (2018) [29] | Filter-guided wavelet fusion | Wavelet/Spatial | Accurate localization of anatomical targets | CT-MRI fusion |
| Saboori & Birjandtalab [30] | Adaptive filtering + spectral-spatial optimization | Spatial | Structural and spectral enhancement through filter parameter tuning | Biomedical instrumentation |
| Pei et al. (2020) [31] | Guided filtering + multiscale layers | Spatial + Texture Layering | Preserved structural layers and enhanced image contrast | Multi-organ medical imaging |
| Tan et al. (2021) [32] | Three-layer image fusion | Spatial Layering | Validated on >100 image pairs from multiple pathologies | Collaborative diagnosis |
| Chen et al. (2021) [33] | Rolling Guidance Filtering | Spatial | Separated and fused structural-detail layers; maintained anatomical clarity | Head and brain imaging |
| Deepali Sale (2022) [34] | PCA, ICA, Averaging | Classical Spatial | Advocated low-complexity spatial fusion for fast implementation | Low-resource medical devices |
| Kong et al. (2022) [35] | Framelet Transform + Subband fusion | Framelet / Spatial | Improved structure clarity via GDGFRW and SWF modules | Radiology, Lesion detection |
| Li et al. (2023) [36] | Modified Laplacian + Local Energy | Spatial Energy-based | Enhanced feature energy preservation; outperformed 9 SOTA methods | Harvard dataset |
| Feng et al. (2023) [37] | SSD for detail and texture retention | Structural Similarity | Solved low contrast and pseudo-edges using structure-preserving fusion | Clinical diagnosis (multi-pathologies) |
| Zhang et al. (2023) [38] | Quasi-cross bilateral filtering (QBF) | Spatial Filtering | Focused on edge contour, lesion detail, and contrast; high benchmark performance. | PET/MRI fusion, Neurology |
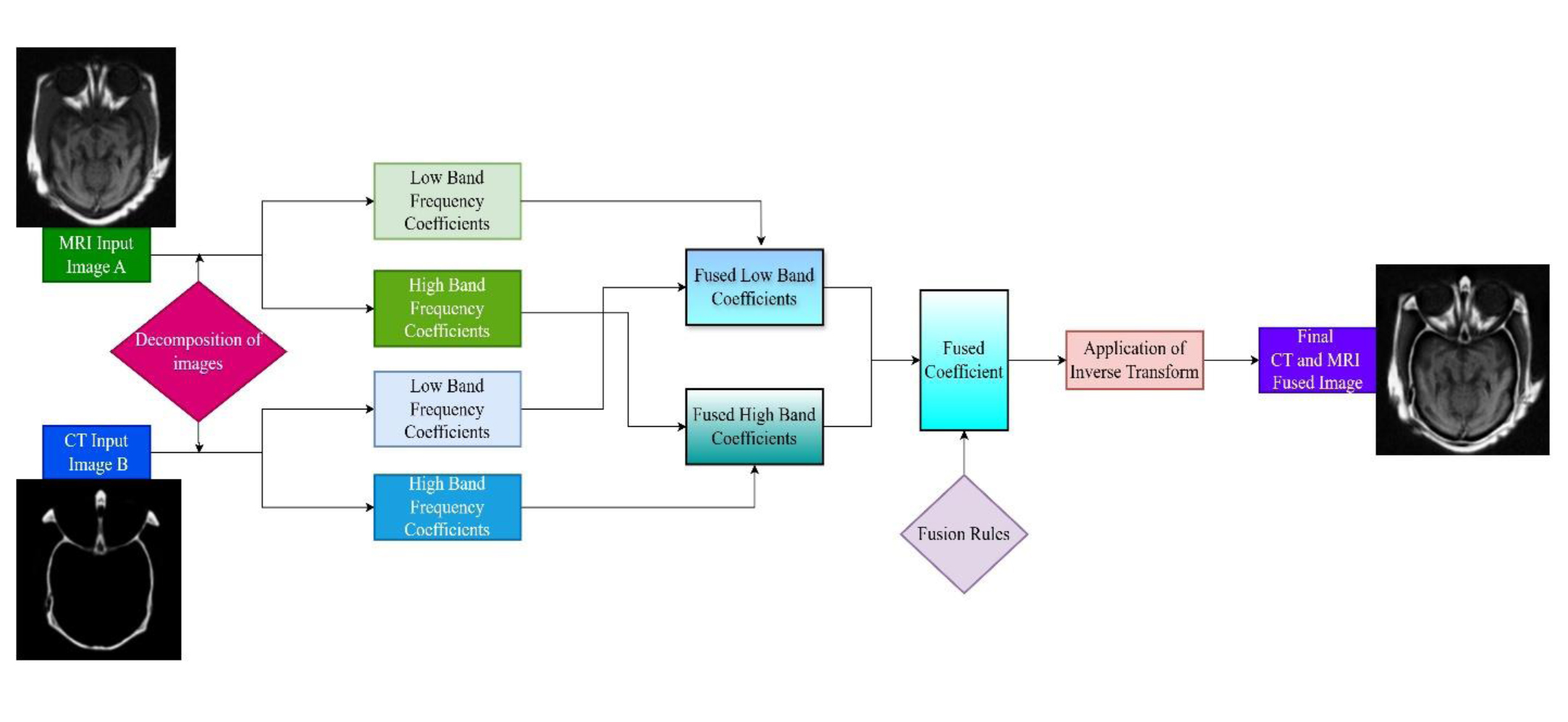
Basic methodology of CT and MRI fusion in the transform domain described along with CT and MRI images from AANLIB [19].
Preliminary research, including that of Singh and Khare [39], tackled the shortcomings of real-valued wavelet transforms, specifically shift sensitivity and inadequate directionality by utilizing the Dual-Tree Complex Wavelet Transform (DCxWT). This method markedly enhanced fusion results by maintaining phase and directional specifics. In the same year, Ganasala and Prasad [40] introduced an image fusion technique for CT and MR images with the Nonsubsampled Contourlet Transform (NSCT), resulting in improved visualization of soft tissue and osseous structures. Bhateja et al. [41] advanced this research by developing a two-stage architecture that included Stationary Wavelet Transform (SWT) and NSCT, utilizing PCA for redundancy minimization and contrast enhancement.
To more effectively capture edge information, Srivastava et al. [42] utilized the Curvelet Transform, which showed enhanced efficacy in maintaining anisotropic features and improving visual perception via a local energy-based fusion rule. Xu et al. [43] presented the Discrete Fractional Wavelet Transform (DFRWT), which, due to its fractional order parameters, facilitated adaptive decomposition and enhanced multimodal image fusion efficacy. Gomathi and Kalaavathi [44] demonstrated that the NSCT method provides improved frequency decomposition while efficiently preserving high-frequency image components. In contrast, Liu et al. [45] showed that the NSST is highly effective in maintaining texture and detail. Numerous scholars investigated shearlet-based and nonsubsampled techniques for improved frequency decomposition. Gambhir and Manchanda [46] also demonstrated that the fused images obtained from the proposed method offer better clarity and enhanced information, making them more useful for quick diagnosis and improved treatment of diseases.
Li et al. [47] improved fusion efficacy by utilizing NSST with a novel fusion rule that reduced blocking and blurring artifacts through local coefficient energy and mean-based methodologies, while Ganasala and Prasad [48] implemented SWT with Transformation Error Minimization (TEM) to improve image fusion quality while decreasing computational load. Goyal et al. [49], Khare et al. [50], and Kong et al. [51] enhanced image fusion by preservation of edges, textures, and structural boundaries, minimizing artifacts with RGF/DTF, median-based NSST, and Framelet Transform, respectively. The results of the work by Diwakar et al. [52] demonstrated that non-conventional transform domains yield improved outcomes when integrated with various spatial domain architectures. An overview of various transform techniques is presented in Fig. (12). Although distinguished by their quality enhancements, frequency domain approaches are accompanied by increased computational costs and high requirements for exact image registration, which may be quite challenging to implement in practice. The key contributions are presented in Table 6.
4.3. Sparse Representation
The sparse representation-based fusion method provides a more compacted, higher information-to-data ratio. Sparse methods examine images by projecting them onto a dictionary of basis functions, which can be trained or predetermined. The focus is on a small set of coefficients that correspond to significant aspects of the image. By combining coefficients from different modalities, as shown in Fig. (13), the method constructs a fused image with enhanced predominant structure and contrast.
| Study | Methodology | Key Contributions |
|---|---|---|
| Singh and Khare [39] | Dual-Tree Complex Wavelet Transform (DCxWT) | Addressed the constraints of wavelet transforms, such as shift sensitivity, poor directionality, for better fusion. |
| Ganasala and Prasad [40] | Nonsubsampled Contourlet Transform (NSCT) | Improved visualization of soft tissue and bone structure, enhancing the quality of image fusion. |
| Bhateja et al. [41] | SWT + NSCT with PCA | Developed a dual-stage architecture for contrast enhancement and redundancy minimization. |
| Srivastava et al. [42] | Curvelet Transform | Effectively captured anisotropic features and improved visual perception with a local energy-based fusion rule. |
| Xu et al. [43] | Discrete Fractional Wavelet Transform (DFRWT) | Delivered adaptive decomposition for enhancing fusion performance across modalities. |
| Gomathi and Kalaavathi [44] | NSCT | Improved frequency decomposition efficiently maintains high-frequency image components. |
| Liu et al. [45] | Nonsubsampled Shearlet Transform (NSST) | Successfully maintain texture and detail effectively. |
| Li et al. [47] | NSST with novel fusion rule | Addressed blocking and blurring artifacts using local coefficient energy and a mean-based fusion technique. |
| Ganasala and Kumar [48] | SWT with Transformation Error Minimization (TEM) | Optimized performance with less computational load and improved image fusion quality. |
| Goyal et al. [49] | Rolling Guidance Filtering (RGF) and Domain Transfer Filtering (DTF) | Maintained edge and texture details while fusing low-resolution images. |
| Khare et al. [50] | Median-based fusion rule within NSST | Preserved structural boundaries through median-based fusion. |
| Kong et al. [51] | Framelet Transform (FT) | Resolved fusion artifacts and texture degradation by decomposing images into structured layers. |
Overview of different techniques of the transform domain.
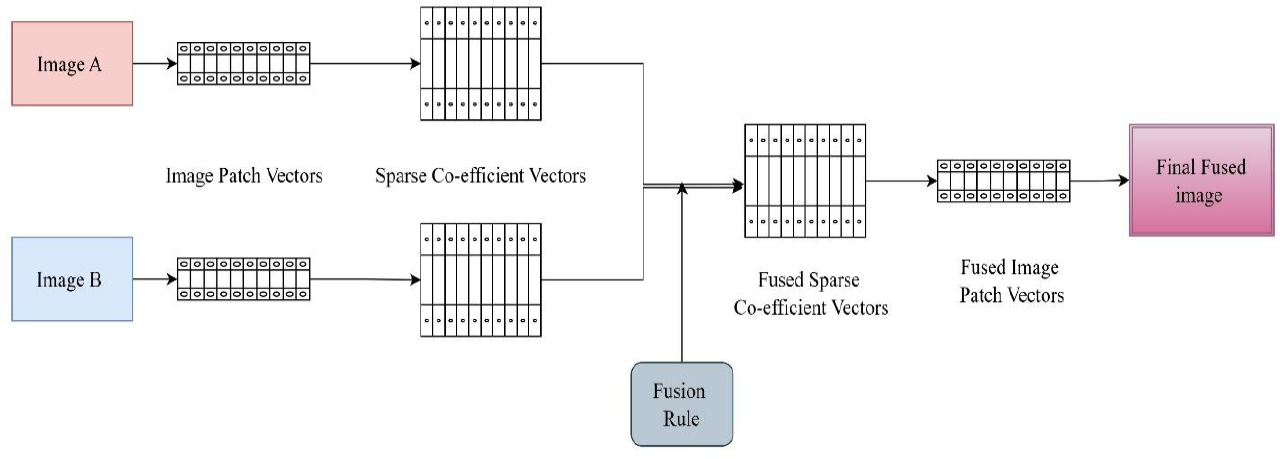
Steps of image fusion using sparse representation methods [53].
It has gained significant interest because of its ability to maintain prominent image elements such as edges and textures while minimizing redundancy. A typical SR-based image fusion process comprises the following steps:
1. Extraction of patches from source images (such as CT and MRI).
2. Encoding of each patch with a trained dictionary.
3. Integration of sparse coefficients by a criterion (e.g., max-selection or averaging).
4. Reconstruction of amalgamated patches to achieve the final image [53].
Zhang et al. [54] described how the SR model forms a dictionary through a sparse linear combination of prototype signal models. According to Joint Sparse Representation (JSR), several signals from several sensors of the same scene constitute an ensemble. While each signal possesses an innovative sparse component, they all share a common sparse component. As compared to SR, the JSR presents reduced complexity. Zong and Qiu [55] proposed a sparse method using categorized image patches based on their geometric orientation. Liu et al. [56] made a significant early contribution by introducing Convolutional Sparse Representation (CSR), an alternate representation of SR using the convolutional form, aiming to achieve SR of a complete image rather than a localized image patch.
Liu et al. [57] presented Convolutional Sparsity-based Morphological Component Analysis (CS-MCA). In contrast to the conventional SR model, which relies on a single image component and overlapping patches, the CS-MCA model can concurrently accomplish multicomponent and SRs of the source images by amalgamating MCA and CSR within a cohesive optimization framework. Shabanzade and Ghassemian [58] employed sparse representation in the NSCT (Nonsubsampled Contourlet Transform) domain in a hybrid context. Their approach integrated low-frequency coefficients through sparse coding and high-frequency components using max-selection, resulting in both multiscale decomposition and sparse adaptability. Alternative hybrid methodologies have integrated SR with PCNN (Pulse Coupled Neural Networks), and clustering-based multi-dictionary learning, which allocates region-specific dictionaries (e.g., for edges versus smooth regions) to enhance localization and context-aware fusion. The various categories of sparse methods are also described in Table 7.
| Category | Model |
|---|---|
| Local and single-component SR-based | Orthogonal Matching Pursuit (OMP), Simultaneous OMP (SOMP), Group Sparse Representation (GSR), Sub-dictionary-based adaptive SR combined with other transforms |
| Multi-component SR-based | Joint Sparse Representation (JSR), Morphological Component Analysis (MCA) |
| Global SR-based | Convolutional Sparse Representation (CSR) |
| Simultaneous multi-component and Global SR-based | MCA-extended version of CSR |
| Aspect | Challenges |
|---|---|
| Feature Preservation [53] | Poor dictionary training can result in suboptimal fusion outcomes. |
| Adaptability [53] | The process of dictionary learning is computationally intensive and requires substantial training data, posing challenges in resource-constrained environments. |
| Robustness to Misregistration [60] | Significant misregistration can still degrade fusion performance, necessitating precise alignment in practice. |
| Computational Complexity [61] | SR methods are computationally demanding, which limits their applicability in scenarios requiring rapid processing. |
| Noise Sensitivity [62] | SR methods may inadvertently amplify noise, compromising image quality if the noise characteristics are not well understood. |
SR continues to serve as a robust intermediary solution between conventional and deep learning-based fusion approaches. Its unsupervised characteristics and adaptability render it especially advantageous in contexts with limited labeled data. As highlighted in the paper by Hermessi et al. [59], SR remains fundamental to numerous advanced approaches, particularly in hybrid fusion architectures where it enhances contrast, clarity, and feature retention [60]. When sparse representation is utilized alongside multi-scale techniques, such as NSCT, Laplacian Pyramid, Dual Tree Complex Wavelet, or Curvelet, it effectively improves fine details, boosts contrast, and minimizes noise. According to a review by Zhang et al. [61], sparse representation proved superior to traditional multi-scale approaches in terms of retaining image structures and defining edges. It learns an overcomplete dictionary from a set of training images, resulting in more stable and significant results. A smart blending approach that combines SR with SCNN to overcome flaws such as edge blurriness, diminished visibility, and blocking artifacts was proposed by Yousif et al. [62]. The results have demonstrated that the proposed method is superior to previous techniques, particularly in suppressing the artifacts produced by traditional SR and SCNN methods. Table 8 summaries the advantages and challenges of this domain. SR-based approaches encounter constraints, including:
- The high computational cost results from sparse optimization procedures.
- Block artifacts arising from independent patch-based judgements.
- Sensitivity to misregistration, as the majority of approaches presume aligned inputs.
4.4. Deep Learning Fusion Methods
The last few years have seen a paradigm shift in MMIF research, with deep learning shaping into an enabler of automated multimodal fusion through end-to-end learning architectures. Although Deep Neural Networks (DNNs) have yielded exceptional results in learning multi-level feature representations from raw image data, the architects of the networks are finding it difficult to give them meaningful names. Notable compositions of DNNs have been CNNs, U-Nets, and Generative Adversarial Networks (GANs). The training objective of these models incorporates optimal structural alignment, maintaining semantic information, and enhancing disease identification.
The initial implementation of CNNs in medical image fusion was presented by Liu et al. [56], exhibiting superior performance as compared to spatial and transform domain approaches. CNNs' design effectively extracts spatial and textural information, yet they necessitate extensive annotated datasets and intricate tuning processes. Liu et al. [63] emphasized the dual roles of fusion rules and activity level estimation, using local filters and clarity maps to guide the amalgamation of high-frequency features. Gibson et al. [64] insisted upon the importance of deep networks for neurological diagnosis through pixel-level fusion.
Xia et al. [65] accomplished a significant advancement by integrating multiscale decomposition with CNNs, facilitating high/low-frequency discrimination and enhanced multi-resolution fusion. Wang et al. [66] employed Siamese CNNs to create activity-guided weight maps, resulting in structurally intricate fused images. To overcome batch processing restrictions, Li et al. [67] created CNN-based frameworks that facilitated real-time, multimodal fusion, therefore improving efficiency and detail retention. These approaches provided practical utility in clinical environments requiring simultaneous processing of many images. The fusion approach using CNN and autoencoders is depicted in Fig. (14).
Chuang et al. [68] introduced a fusion framework that integrated U-Net and Autoencoder architectures (FW-Net), where the encoder-decoder configurations exhibited U-Net’s skip connections. The design, initially limited to CT-MRI, has the potential for future PET-MRI and SPECT fusion. Kumar et al. [69] examined the utilization of CNNs for thermal-visual fusion in remote sensing, showing cross-domain relevance. El-Shafai et al. [70] introduced a hybrid fusion technique in which CNNs integrated three images using traditional methods, leading to less redundancy and enhanced semantics.
To address the issues of semantic degradation in fused outputs, Ghosh [71] introduced a dual U-Net FW-Network that prioritized semantic-level fusion. This approach markedly enhanced clinical utility in disease localization and segmentation. Deep learning-based image fusion is a powerful paradigm that provides excellent visual fidelity, task adaptability, and end-to-end automation. However, accessible data sets, interpretable models, and the creation of computationally efficient architectures that work across modalities are necessary for the advancement.
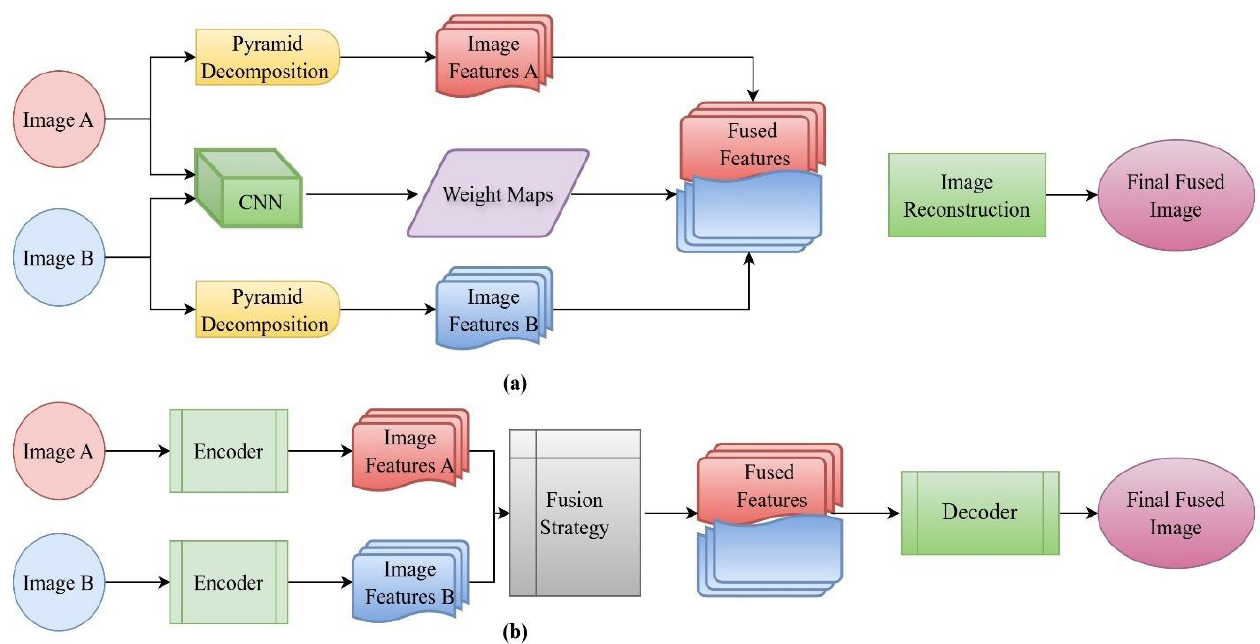
Different approaches of deep learning (a) CNN-based fusion (b) Autoencoder-based fusion [2].
A CNN-based multimodal fusion approach designed for oncology not only succeeded in improving diagnostic accuracy but also outperformed current methods in both subjective and objective metrics. Due to their generative properties, GANs can create high-resolution fused images that preserve both anatomical structure and functional intensity. The dependence of such models on large sets of labeled data and their tendency to overfit limited data variability are serious issues. The lack of interpretability is also a concern for these models in clinical settings. Figure 15 demonstrates the domain-wise publication trends across Web of Science, in multimodal image fusion from 2000 to 2024. It indicates the rapid proliferation of deep learning and hybrid domains in this period. Driven by the endorsement of CNNs, U-Nets, and GANs, the adoption of deep learning methodologies has shown remarkable expansion. The explosion in growth indicates a shift in the sphere towards using data-driven fusion techniques, characterized by a strong understanding and fusion of intricate anatomical and functional relationships.
At the same time, the development of hybrid approaches using such spatial, frequency, and deep learning approaches is relatively stable, demonstrating their ability to handle the dissimilar nature of medical data. Due to their modular structure, these techniques provide superior flexibility and precision, which improves overall fusion performance. Collectively formed by these advances, these fields have outperformed traditional approaches and indicate a shift towards smarter, more holistic fusion methods for medical applications. MMIF's future research appears to be majorly focused on DL-based and hybrid techniques, especially where complex cases of oncology and neuroimaging are involved. The key contributions from DL are presented in Table 9.
4.5. Hybrid Domain
To benefit from diverse domains and tend towards their minimal negative effects, hybrid fusion approaches have attracted much attention. Such approaches utilize specific domain expertise, such as NSCT for frequency analysis and CNNs for feature extraction, or the use of neural networks. Simultaneously managing the spatial, frequency, and contextual information, hybrid systems are capable of creating informative fused images.
Xia et al. [72] established a foundational framework by combining Nonsubsampled Contourlet Transform (NSCT) and Dual-Tree Complex Wavelet Transform (DTCWT), which was supplemented by Pulse-Coupled Neural Networks (PCNN) for the creation of composite images. This represented a crucial advancement in integrating handcrafted and adaptive transformations to tackle challenges such as image noise and resolution discrepancies. Wang et al. [73] improved diagnostic precision by employing NSST and adaptive decomposition frameworks that consider high/low frequency layers and dynamic textural features, respectively. These works acknowledged that conventional static decomposition inadequately reflects contextual differences across modalities. Bhateja et al. [74] advanced the hybrid approach by introducing shearlet and NSCT-based fusion for PET/SPECT, MRI, and CT images, including contrast enhancement and weighted PCA to preserve multispectral integrity. Du et al. [75] and Zhao et al. [76] tackled modality-specific inconsistencies by developing distinct methodologies for MRI and PET, thereby enhancing fusion accuracy through edge-based weighting.
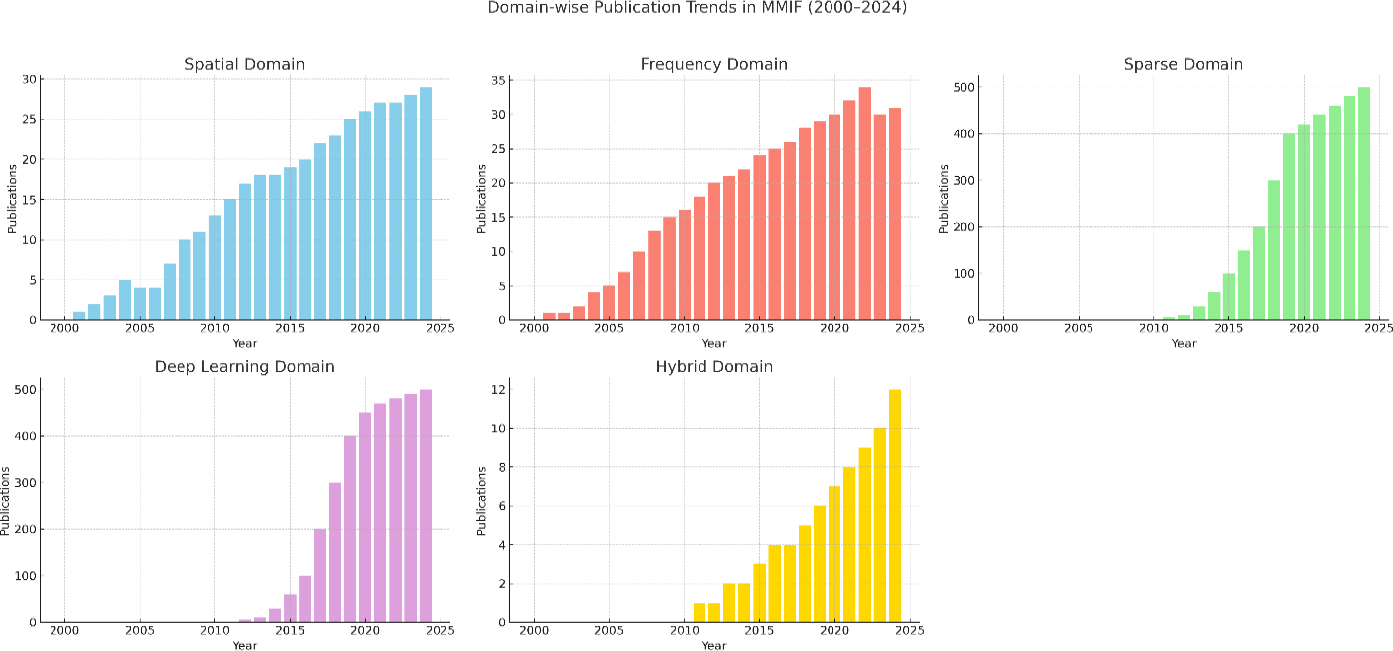
Publication trends across all domains over the last five years (WOS).
| Study | Methodology | Key Contributions |
|---|---|---|
| Liu et al. [56] | Convolutional Neural Networks (CNNs) | Exhibited better performance as compared to spatial and transform domain methods, highlighting the capability of CNN to extract spatial and textural characteristics. |
| Liu et al. [63] | CNNs with fusion rules and activity level estimation | Implemented local filters and clarity maps to facilitate the integration of high-frequency features, enhancing fusion quality. |
| Gibson et al. [64] | Deep networks | Highlighted the significance of deep learning in neurological diagnosis and enhancing fusion at the pixel level for diagnostic accuracy. |
| Xia et al. [65] | Multiscale decomposition combined with CNNs | Better division of high/low-frequency components, facilitating detailed and multi-resolution fusion. |
| Wang et al. [66] | Siamese CNNs with activity-guided weight maps | Increased structural richness in final fused images via activity-guided weight maps. Enhancing the retention of essential features |
| Li et al. [67] | CNN-based | Concentrated on real-time processing of multimodal images, providing practical utility in clinical settings. |
| Chuang et al. [68] | Fusion U-Net and Autoencoder (FW-Net) | A hybrid encoder-decoder structure was introduced, with potential for further expansion to PET-MRI and SPECT fusion. |
| Kumar et al. [69] | CNNs | Proposed cross-domain applications, using CNNs for thermal-visual fusion. |
| El-Shafai et al. [70] | Hybrid CNN-based fusion pipeline | Combined three images using traditional approaches, providing a reduction in redundancy and enhancing the semantic quality in fused outputs. |
| Ghosh [71] | Dual U-Net FW-Network | Concentrated on enhancing disease localization and segmentation by addressing semantic degradation in fused outputs. |
Maqsood and Javed [77] employed two-scale decomposition with spatial gradients to enhance edges. Wang et al. [78] and Liu et al. [79] employed adaptive sparse coding and total variation transformations, enhancing detail retention while diminishing high-frequency noise. Yadav [80] utilized independent and principal component analysis using a wavelet framework but observed residual noise and artifacts. In their study, Ashwanth and Swamy [81] showed that the Stationary Wavelet Transform (SWT) surpasses the standard Discrete Wavelet Transform (DWT) in terms of entropy preservation. Huang et al. [82] emphasized that while hybrid models enhance established frameworks, problems persist, especially in feature extraction and color distortion. These constraints stimulated investigation into hybrid deep learning strategies. Kaur et al. [83] integrated ANFIS with cross-bilateral filtering, resulting in enhanced entropy (2.92), and suggested volumetric fusion for future neuroimaging applications. Kaur et al. [84] conducted a systematic review emphasizing the necessity for precise, reliable, and interpretable fusion techniques.
Polinati et al. [85] and Li et al. in 2021 [86] introduced innovative frameworks employing Variational Mode Decomposition (VMD), local energy gradients, and bilateral filtering, all designed to improve clarity and reduce luminance deterioration. Zhu et al. [87] and Alseelawi and Hazim [88] concentrated on enhancing NSST-DTCWT-based fusion techniques to achieve equilibrium between texture and structural integrity. Alseelawi’s work prominently highlighted velocity and visual excellence, using PCNN as a guiding framework. Kalamkar and Geetha [89] utilized transfer learning with DWT, attaining enhanced structural similarity index values. Goyal et al. [90] developed a cross-bilateral, edge-preserving fusion filter that reduces artifacts while improving image quality. Likewise, Faragallah et al. [91] amalgamated CNN with three traditionally fused image sets to improve overall image quality and diagnostic efficacy. Zhou et al. [92] tackled issues of brightness degradation and detail retrieval by the application of NSST and Improved Structure Tensor (IST) decomposition, resulting in enhanced contrast while differentiating smoothing, edge, and corner layers.
Kittusamy and Kumar et al. [93] utilized Joint Sparse Representation (JSR) and NSCT to attain enhanced high perceptual clarity in MRI-CT fusion. Dinh [94, 95] formulated two complementary models to address inadequate contrast and edge degradation through a three-scale decomposition and local energy-based fusion rules. Balakrishna et al. [96] evaluated nine DWT-based combinations, validating the method’s efficacy in the detection of abnormalities and clinical planning. Moghtaderi et al. [97] suggested a Multilevel Guided Edge-Preserving Filtering (MLGEPF) technique that combined computational expense with structural accuracy. Zhao et al. [98] proposed a novel method based on three-scale frequency decomposition along with SSIM-optimized feature blending, which significantly facilitates the process of fusing MRI and PET images for brain tumor examination. Various hybrid combinations, along with their advantages and disadvantages, are described in Table 10.
Table 11 signifies the evolving emphasis of research within Multimodal Medical Image Fusion (MMIF) over three distinct phases. Spatial domain methods prevailed in the initial period but have since been replaced by more sophisticated techniques. Frequency domain techniques have shown significant output growth and have sustained stability. Sparse representation techniques have transitioned from minimal utilization in their first stages to considerable significance in recent years.
| Author/ Year | Hybrid Combination | Advantages | Disadvantages |
|---|---|---|---|
| Xia et al., 2018 [72] | NSCT + PCNN | Improves contrast; effective for multi-resolution fusion | Complex; sensitive to noise levels and parameter tuning |
| Wang et al., 2018 [73] | Adaptive Decomposition + Texture Integration | Dynamic layer integration improves color and texture representation | Requires rule tuning; may introduce artifacts |
| Bhateja et al., 2018 [74] | Shearlet + NSCT + Weighted PCA | High spectral-spatial fidelity; suited for PET/CT/MRI | Computationally intensive; color normalization required |
| Du et al., 2019 [75] | Separate Decomposition for MRI and PET | Customized decomposition maintains modality-specific features | High design complexity; lacks real-time scalability |
| Wang et al., 2020 [78] | Adaptive Sparse + Laplacian Pyramid | Minimizes noise in high-frequency ranges; adaptive sparsity | May be unstable with varying image types |
| Yadav and Yadav, 2020 [80] | Wavelet + ICA/PCA | Elementary; broadly applicable; improves interpretability | Noise sensitivity; potential overfitting |
| Ashwanth and Swamy, 2020 [81] | SWT vs DWT | SWT retains greater energy and detail compared to DWT | Requires precise coefficient tuning |
| Kaur et al., 2021 [83] | ANFIS + Cross-Bilateral Filter | Enhanced entropy; edge detail maintained | Requires ANFIS model training and tuning |
| Faragallah et al., 2022 [91] | CNN + Multi-Input Fusion | Efficient and effective; reduces redundancy | Quality depends on pretrained models |
| Kalamkar and Geetha, 2022 [89] | Transfer Learning + DWT | Improved structural similarity and generalization | Training requires large datasets |
| Zhou et al., 2022 [92] | NSST + IST (Improved Structure Tensor) | Maintains brightness and local structure | Complex to implement; computationally heavy. |
| Kittusamy and Kumar, 2023 [93] | JSR + NSCT | Elevated contrast and detail retention in soft tissues | Requires dictionary learning; time-consuming |
| Dinh, 2023 [94] | Three-Scale Decomposition + Local Energy | Prevents information loss in edges; enhances contrast | Parameter tuning is crucial; risk of blur |
| Moghtaderi et al., 2024 [97] | Guided Edge-Preserving Filtering (MLGEPF) | Prompt and reliable; balances performance and quality | Filter design and scale sensitivity |
| Zhao et al., 2024 [98] | Guided Fusion (Smoothing + Global Optimization) | Maintains texture, noise, and structure clearly | May overlook fine details if the structure is misclassified |
| Domain | Early Phase (2000–2010) | Growth Phase (2011–2017) | Boom Phase (2018–2024) |
|---|---|---|---|
| Spatial | High | Medium | Declining |
| Frequency | Medium | High | Stable |
| Sparse Representation | Very Low | Medium | High |
| Deep Learning | None | Emerging (post-2016) | Dominant |
| Hybrid | Low | Emerging | Rising sharply |
| Domain | Advantages | Limitations | Typical Applications |
|---|---|---|---|
| Spatial Domain [19] |
Elementary execution, Better color representation. Rapid computation | Edge blurring, Low SNR, Spectral distortion | SPECT-MRI fusion using HSV- X-ray/CT overlays |
| Frequency Domain [50] | Elevated SSIM- Good texture and edge preservation | Complex registration, High computational cost | Tumor localization (MRI-PET)- Brain mapping using NSCT and Laplacian Pyramid |
| Sparse Representation [60, 61] | Enhanced contrast clarity-Concise representation, Good noise removal | Artifacts from basis mismatch, Edge degradation | CT-MRI brain fusion utilizing K-SVD- Dictionary learning for tumor detection |
| Deep Learning [4, 64] |
Automatically acquires complex features, High fusion accuracy | Requires large datasets, Risk of overfitting | U-Net fusion in COVID detection- GANs for whole-body PET/CT integration |
| Hybrid Domain [83, 88] |
Combines domain strengths, High robustness | Increased model complexity, Difficult tuning | NSCT + PCNN fusion- DWT-IFS-PCA in multimodal cancer detection |
Deep learning was nearly non-existent before 2016, but has become the dominant methodology. Hybrid approaches, which amalgamate multiple techniques, have experienced significant expansion, signifying a trend towards integrative and adaptive fusion approaches. A comprehensive analysis of Multimodal Medical Image Fusion (MMIF) domains emphasizing their strengths, weaknesses, and principal application areas is highlighted in Table 12.
5. APPLICATIONS OF IMAGE FUSION IN CLINICAL MEDICINE
This section highlights the integration of structural medical imaging modalities through fusion techniques and their clinical applications, especially in surgical planning, oncology, orthopedics, and neurosurgery. The following examples highlight recent advances and use cases where image fusion has improved diagnosis, treatment accuracy, and procedural efficiency.
5.1. Surgical Planning Using Synthetic CT from MRI
A recent study has explored the feasibility of generating synthetic Computed Tomography (CT) images of the lumbar spine from Magnetic Resonance Imaging (MRI) using a patch-based convolutional neural network. This approach aimed to support pre-operative planning without exposing patients to ionizing radiation using deep learning. The study involved three cases and demonstrated that deep learning-enabled MRI-to-CT conversion offers high-quality structural visualization, reducing radiation exposure compared to conventional CT scans (typically 3.5–19.5 mSv for spine imaging) [99].
5.2. Orthopedic Implant Design and Additive Manufacturing
The combination of CT and MRI has also been pivotal in the design of patient-specific orthopedic implants. These modalities are employed to capture comprehensive anatomical data, including size, shape, texture, and bone density. This facilitates the development of custom implants and allows for the reconstruction of traumatic bone defects through additive manufacturing technologies [100].
5.3. Management of Complex Fractures Using Rapid Prototyping
Fusion imaging has enabled advances in Rapid Prototyping (RP) and 3D reconstruction, particularly for complex fractures in anatomical regions such as the joints, acetabulum, and spine. RP assists surgeons in visualizing fracture geometry preoperatively, which improves the accuracy of anatomical reduction and reduces surgical time, anesthesia duration, and intraoperative blood loss [101].
5.4. Surgical Effects of Resecting Skull Base Tumors Using Preoperative Multimodal Image Fusion
A retrospective study was conducted on 47 patients with skull base tumors. Preoperative CT and MRI data acquisition were performed using GE AW workstation software for co-registration, fusion, and three-dimensional reconstruction of the brain. The surgical plan was designed based on multimodal images. The application of the fusion technique provided essential visual guidance in skull base tumor surgery, assisting neurosurgeons in accurately planning the surgical incision and precisely resecting the lesion [102].
5.5. Trigeminal Neuralgia Treatment with Integrated Neuro-Navigation
In 13 patients undergoing percutaneous radiofrequency trigeminal rhizotomy, the fusion of MRI and intraoperative CT (iCT) images provided improved anatomical delineation of the trigeminal cistern. This integration supports more accurate targeting, especially in recurrent cases of trigeminal neuralgia. The study emphasized the benefits of fusion-guided neuro-navigation and suggested that longer follow-ups are needed to assess long-term therapeutic efficacy [103].
5.6. Frameless Stereotactic Radiosurgery with Gamma Knife Icon
In a clinical series of 100 patients, MRI-CBCT (Cone Beam CT) fusion was utilized for frameless stereotactic radiosurgery using the Gamma Knife Icon. MRI provided superior soft tissue contrast, while CBCT served as the baseline for stereotactic registration. The adaptive dose distribution was computed based on the patient's real-time geometry after fusion, optimizing treatment accuracy. This method overcomes the limitations of traditional CT-guided radiotherapy by ensuring better alignment of tumor and anatomical landmarks [104, 105].
5.7. Image Fusion in Precision Medicine for Oncology
In the context of precision oncology, fusion imaging technologies are increasingly used to enhance diagnostic accuracy and individualized treatment planning. The integration of CT, MRI, and PET enables better visualization of tumor morphology and metabolic activity, facilitating more accurate localization and classification of malignancies. Such multimodal fusion approaches are pivotal in improving therapeutic outcomes and reducing harm to healthy tissue [106].
5.8. Image Fusion in the Diagnosis and Treatment of Liver Cancer
The rapid advancement of medical imaging has facilitated the effective application of image fusion technology in diagnosis, biopsy, and radiofrequency ablation, particularly for liver tumors. Employing image fusion technology enables the acquisition of real-time anatomical images overlayed with functional images of the same plane, thereby enhancing the diagnosis and treatment of liver cancers. This study provides a comprehensive examination of the fundamental concepts of image fusion technology, its application in tumor therapies, specifically for liver cancers. It finishes with an analysis of the limitations and future prospects of this technology [107].
6. IMAGE QUALITY METRICS IN MEDICAL IMAGE FUSION
The assessment of medical image fusion is crucial for verifying the efficacy of fusion algorithms. Image quality measures evaluate the visual integrity, information retention, and diagnostic value of the fused image. These assessments are conducted utilizing either subjective or objective approaches [108, 109].
6.1. Subjective Evaluation
Subjective analysis entails expert evaluators who assess the images according to their visual characteristics, such as object clarity, spatial detail, geometric consistency, and color equilibrium. This method, although indicative of human perception, is compromised by observer bias, environmental reliance, and lack of reproducibility, rendering it less reliable for quantitative comparison.
6.2. Objective Evaluation
It utilizes the mathematical and statistical metrics to measure image quality quantitatively. These metrics yield reliable, quantifiable, and algorithm-independent outcomes, making them crucial for the consistent and automated validation of fusion approaches. They are further categorized into two categories such as those employing a reference image and those without a reference image, as depicted in Table 13, with each parameter demonstrating distinct characteristics. The objective image fusion performance characterization utilizing the gradient information is also taken into account. This provides an in-depth analysis by assessing total fusion performance, fusion loss, and fusion artifacts as represented in Table 14. It is noted that the total fusion performance is depicted by the sum of these three, and the result is unity, as shown in the formula [110-113].
7. EXPERIMENTAL SETUP AND DISCUSSION
In this section, a comprehensive evaluation of eight multimodal medical image fusion techniques, LEGFF, FGF-XDOG, MDHU, FDO-DPGF, CSMCA, S-ADE, PCLLE-NSCT, and NSST-AGPCNN, is conducted for dataset 1 [114] utilizing diverse quantitative criteria to evaluate information content, image quality, edge preservation, and noise reduction, and has been described in Table 15 along with mean and standard deviation. Of these evaluated approaches, LEGFF exhibited the highest entropy of 6.86 and average gradient of 7.06, signifying enhanced information richness and edge definition, while the mean entropy value is 6.44 ± 0.35. CSMCA demonstrated superior image quality, attaining the highest PSNR (63.28) and the lowest MSE (0.0305), indicative of exceptional image reconstruction with minimum error. The spatial frequency values, which assess image detail, showed a moderate spread (17.79 ± 0.56), demonstrating that CSMCA and LEGFF retained excellent textural detail. In contrast, standard deviation results have revealed that LEGFF and S-ADE maintained a significant contrast. FGF-XDOG and S-ADE achieved the best visual information fidelity (VIF ~0.88), indicating superior perceptual quality. PCLLE-NSCT exhibited superior structural similarity, achieving a reduced spatial correlation difference (SCD = 1.72) compared to others. The Correlation Coefficient (CC) analysis indicated a preference for CSMCA and FDO-DPGF (CC ~0.698), emphasizing their robust correspondence with reference images.
| Metric Name | Description | Formula / Expression | - |
|---|---|---|---|
| Average Pixel Intensity (API) | Measures image contrast by computing average intensity values. |
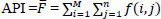
|
[110, 111] |
| Standard Deviation (SD) | Measures the contrast or spread of the pixel intensity values around the mean intensity. A higher SD indicates more variation and, therefore, more detail and contrast. |
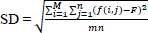
|
[112] |
| Average Gradient (AG) | Evaluates the overall clarity and detail of the image by computing the average magnitude of the gradients in the image. To assess the sharpness and clarity of an image and quantify the overall contrast by measuring the rate of intensity change across adjacent pixels. |
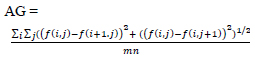
|
[112] |
| Entropy (H) | Quantifies information content or randomness in the image. A higher entropy value indicates a richer and more complex image. |
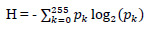
|
[113] |
| Mutual Information (MI) | Assesses shared information between source and fused images. It should have a high value for better fusion. |
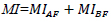
|
[110, 113] |
| Information Symmetry (FS) | Evaluates the symmetry of information between fused and input images. |
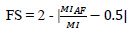
|
[110] |
| Correlation Coefficient (CC) | It gives similarity in the small structures between the original and reconstructed images, where a higher value of correlation means more information is preserved. Determines the linear relationship between input and fused images. |
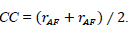
|
[111] |
| Spatial Frequency (SF) | Measures the overall activity level or texture of an image, combining the row and column frequency components. |
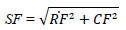
|
[113] |
| Metric Name | Description | Formula / Expression | Citations |
|---|---|---|---|
| Qab/f | Quantifies retained information from the source to the fused image. | Qab/f +Lab/f + Nab/f =1 | [110, 112] |
| Lab/f | Measures information loss during fusion. | Qab/f +Lab/f + Nab/f =1 | [110, 112] |
| Nab/f | Estimates artifacts or noise introduced after fusion. | Qab/f +Lab/f + Nab/f =1 | [110, 112] |
Technique

|
LEGFF | FGF-XDOG | MDHU | FDO-DPGF | CSMCA | S-ADE | PCLLE-NSCT | NSST -AGPCNN | Mean± SD |
|---|---|---|---|---|---|---|---|---|---|
Evaluation Parameter
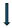
|
|||||||||
| EN | 6.857 | 6.407 | 5.992 | 6.789 | 6.378 | 6.005 | 6.803 | 6.296 | 6.44 ± 0.35 |
| SF | 18.72 | 17.37 | 18.32 | 17.36 | 18.13 | 17.85 | 17.16 | 17.36 | 17.79 ± 0.56 |
| SD | 59.48 | 59.74 | 59.38 | 53.64 | 51.91 | 58.99 | 57.39 | 49.40 | 56.24± 4.03 |
| PSNR | 61.85 | 61.55 | 61.53 | 62.72 | 63.27 | 61.58 | 61.93 | 63.55 | 62.26 ± 0.82 |
| MSE | 0.0424 | 0.0454 | 0.0456 | 0.0347 | 0.0305 | 0.0451 | 0.0416 | 0.0321 | 0.04 ± 0.01 |
| MI | 3.046 | 4.710 | 5.854 | 3.074 | 2.328 | 5.795 | 3.291 | 1.489 | 3.70 ± 1.60 |
| VIF | 0.776 | 0.885 | 0.874 | 0.869 | 0.652 | 0.884 | 0.797 | 0.483 | 0.78 ± 0.14 |
| AG | 7.059 | 6.440 | 6.511 | 6.332 | 6.574 | 6.460 | 6.551 | 6.182 | 6.51 ± 0.25 |
| CC | 0.671 | 0.686 | 0.666 | 0.698 | 0.698 | 0.663 | 0.652 | 0.645 | 0.67 ± 0.02 |
| SCD | 1.823 | 1.898 | 1.841 | 1.766 | 1.746 | 1.831 | 1.724 | 1.453 | 1.76 ± 0.14 |
| Qabf | 0.731 | 0.774 | 0.778 | 0.771 | 0.654 | 0.776 | 0.739 | 0.600 | 0.73 ± 0.07 |
| Nabf | 0.018 | 0.021 | 0.024 | 0.012 | 0.020 | 0.027 | 0.025 | 0.018 | 0.02 ± 0.00 |
FGF-XDOG and MDHU attained the highest edge-based similarity (Qabf >0.77), indicating efficient edge retention, which is essential in diagnostic imaging. FDO-DPGF demonstrated the lowest noise-based similarity (Nabf = 0.0126), indicating exceptional noise suppression. Mutual Information (MI), crucial for evaluating modality complementarity, showed significant variance (mean = 3.7 ± 1.5), with MDHU and S-ADE outperforming. Visual Information Fidelity (VIF) and Qabf. NSST-AGPCNN and LEGFF regularly score well across most criteria, demonstrating superior fusion quality. Future studies should provide larger datasets with repeated trials to allow for more rigorous statistical inferences.
The visual results in Fig. (16) also depict similar results where the source image (a) is a T1-weighted MRI image characterized by the dark appearance of cerebrospinal fluid and the high contrast between gray and white matter. CSF appears hypointense owing to the diminished signals on T1 T1-weighted sequence. T1-weighted scans offer superior anatomical detail and are commonly employed to evaluate brain morphology and the integrity of cerebral structures. The source image (b) is a non-contrast computed tomography scan of the brain, highlighting the bony content. CT scans are highly effective in identifying acute bleed, cerebral infarcts, fractures, and calcifications owing to their sensitivity to dense tissues such as bone. Bone exhibits hyper-density (white) because of elevated X-ray attenuation as compared to soft tissue. The images from (c) to (j) are the final fused images of these source images using the respective technique.
Table 16 provides an evaluation for dataset 2 [114], using the same set of techniques, and demonstrates diverse efficacy across essential quantitative parameters. The average Entropy (EN) is 5.30 ± 0.09, reflecting consistent information content with minimal variance. Spatial Frequency (SF), which is an indicator of textural detail, showed greater variation (20.38 ± 1.66), with LEGFF achieving the highest score (23.77), suggesting better edge detail retention.
Standard Deviation (SD) and PSNR were also stable, with PSNR averaging 68.44 ± 0.52 dB, indicating excellent noise suppression and fidelity. The Mean Square Error (MSE) values remained low (0.01 ± 0.00), reinforcing the high-quality reconstruction by all techniques, particularly NSST-AGPCNN (lowest MSE = 0.0074).
Mutual Information (MI) values showed moderate variance (3.87 ± 0.61), with S-ADE standing out as having the highest MI (4.976), highlighting its capacity to retain complementary modality features. VIF and CC metrics also revealed strong visual and structural correlation, particularly for FDO-DPGF and LEGFF. All techniques performed well with minor variability across metrics. Techniques like NSST-AGPCNN and S-ADE demonstrated superior balance in contrast enhancement, edge retention, and structural fidelity.
Qualitative results for MRI/CT images. Dataset 1 (a) MRI image, (b) CT image, (c) LEGFF, (d) FGF-XDoG, (e) MDHU, (f) FDO-DPGF, (g) CSMCA, (h) S-ADE, (i) PCLLE NSCT, (j) NSST AGPCNN.
Table 16.
Technique

|
LEGFF | FGF-XDOG | MDHU | FDO-DPGF | CSMCA | S-ADE | PCLLE-NSCT | NSST -AGPCNN | Mean ± SD |
|---|---|---|---|---|---|---|---|---|---|
Evaluation Parameter
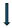
|
|||||||||
| EN | 5.336 | 5.322 | 5.210 | 5.262 | 5.426 | 5.317 | 5.382 | 5.161 | 5.30 ± 0.09 |
| SF | 23.77 | 20.43 | 20.40 | 18.12 | 20.20 | 20.13 | 18.91 | 21.07 | 20.38 ± 1.66 |
| SD | 61.97 | 62.11 | 61.12 | 57.59 | 56.33 | 61.59 | 58.77 | 55.31 | 59.35 ± 2.71 |
| PSNR | 68.36 | 68.26 | 68.01 | 68.24 | 68.82 | 67.76 | 68.63 | 69.42 | 68.44 ± 0.52 |
| MSE | 0.0094 | 0.0096 | 0.0102 | 0.0097 | 0.0085 | 0.0108 | 0.0088 | 0.0074 | 0.01 ± 0.00 |
| MI | 3.426 | 4.088 | 4.378 | 4.118 | 3.233 | 4.976 | 3.618 | 3.132 | 3.87 ± 0.63 |
| VIF | 0.577 | 0.594 | 0.625 | 0.870 | 0.528 | 0.682 | 0.625 | 0.517 | 0.63 ± 0.11 |
| AG | 6.397 | 5.580 | 5.212 | 4.723 | 5.754 | 5.355 | 5.348 | 5.681 | 5.51± 0.48 |
| CC | 0.926 | 0.927 | 0.914 | 0.903 | 0.914 | 0.910 | 0.919 | 0.923 | 0.92 ± 0.01 |
| SCD | 1.249 | 1.268 | 1.106 | 0.730 | 0.769 | 1.114 | 0.992 | 0.790 | 1.00 ± 0.22 |
| Qabf | 0.595 | 0.609 | 0.605 | 0.609 | 0.546 | 0.645 | 0.585 | 0.583 | 0.60 ± 0.03 |
| Nabf | 0.017 | 0.027 | 0.018 | 0.0153 | 0.028 | 0.006 | 0.035 | 0.0203 | 0.02 ± 0.01 |
Qualitative results for MRI/CT images Date set 2 (a) CT image, (b) MRI image, (c) LEGFF, (d) FGF-XDoG, (e) MDHU, (f) FDO-DPGF, (g) CSMCA, (h) S-ADE, (i) PCLLE NSCT, (j) NSST AGPCNN.
The visual results for dataset 2 are depicted in Fig. (17), where the source image (a) is a computed tomography scan of the brain proficient in assessing intracranial bleed and cerebral fractures. The image provides a clear depiction of the bone architecture with no evident indications of structural abnormalities. The source image (b) is a T2-weighted MRI scan of the brain, illustrating the differentiation of gray and White matter, and is marked by bright cerebrospinal fluid. The imaging modality is effective for identifying pathological alterations, including edema, demyelination, and infarctions. The images from (c) to (j) are the final fused images of these source images using the respective technique.
7.1. Research Trends in Modality Integration
The growing interest in MMIF studies from 2015 to 2024, analyzed according to the three most commonly used modality groups, MRI–PET, MRI–CT, and MRI–SPECT, is illustrated in Fig. (18). MRI–PET fusions report the greatest number of publications, with a significant boost in 2021, representing high influence in oncology and neuroimaging. The constant growth of MRI–CT literature (although its volume is less compared to MRI–PET) indicates growing use for surgical planning, and tasks where both soft and hard tissue information is needed.
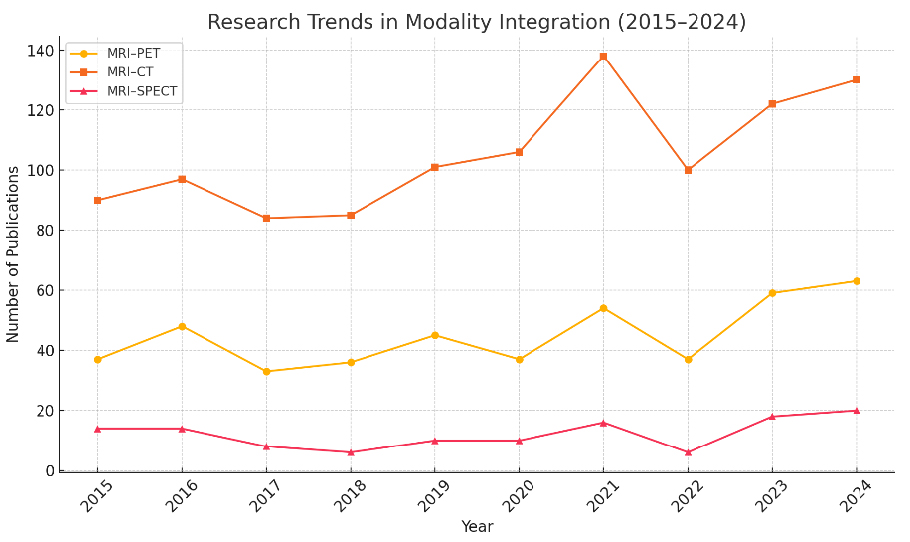
Research Trend according to multi-modality combinations (WOS).
The MRI–SPECT fusion approach is the least discussed, possibly due to the lower spatial resolution and the comparatively narrower use of SPECT compared to PET. The possible correlation of periodic changes in MRI–PET and MRI–CT domains with the emergence of new advances, such as convolutional neural networks or transformer models, indicates that these improvements have likely reinforced the fusion and registration processes. The data shows an increasing interest in hybrid imaging as the main research topic, highlighting the clinical necessity for accurate diagnostics based on the integration of complementary modalities.
8. KEY CHALLENGES AND LIMITATIONS
8.1. Data Scarcity and Imbalance in Public Datasets
The foundation of any robust MMIF model is a sufficiently large and diverse dataset. However, available multimodal image datasets such as AANLIB, ADNI, TCIA, and MIDAS lack balance in organ representation, modality pairing, and demographic diversity. While these repositories provide aligned pairs (e.g., MRI–PET), they often suffer from incomplete labeling and variation in acquisition protocols. For example, Venkatesan et al. [15] emphasized that most fusion research focuses on brain datasets (MRI-CT), leaving thoracic, abdominal, and musculoskeletal fusions underrepresented. The imbalance leads to biased models that cannot generalize across anatomical regions. Moreover, annotating multimodal images, particularly PET and SPECT, requires domain expertise and is cost-intensive, limiting supervised learning approaches. Recent works, such as those by Tirupal et al. [8], propose the use of fuzzy sets and unsupervised learning to overcome the lack of labels, while Dinh [94, 95] explores decomposition techniques to create proxy supervision.
8.2. Registration and Inter-modality Inconsistency
Accurate registration is pivotal in MMIF, as misalignment between modalities can propagate errors into every subsequent fusion stage. Unlike unimodal registration, where intensity similarities guide optimization, multimodal images exhibit diverse characteristics (e.g., CT for density, MRI for soft tissues, PET for metabolism), making intensity-based alignment ineffective. Errors in registration introduce artifacts, structural shifts, and contrast mismatches, especially at organ boundaries. Goyal et al. [90] and Ibrahim et al. [109] report that deformable and affine transformations often underperform when the inter-modality gap is large, such as in PET–MRI or SPECT–CT fusion. Furthermore, hybrid methods that employ wavelet or Laplacian transforms are highly sensitive to minor misregistrations, causing blur or duplication of anatomical features. Although deep-learning-based spatial transformers, such as the Swin Transformer by Ghosh [71], offer improved alignment, these solutions are computationally expensive and poorly validated across multiple organs. Thus, the modality inconsistency and lack of robust, generalized registration algorithms significantly hinder the clinical reliability of MMIF systems.
8.3. Computational Overhead in Deep Learning-based MMIF
While Deep Learning (DL) methods, particularly CNNs, GANs, and U-Nets, have revolutionized image fusion, they bring substantial computational burdens. Training these models requires massive datasets, high-end GPUs, and time-intensive tuning. Liu et al. and Zhang et al. [60, 61] showed that deep CNNs produce sharper and semantically richer fused images, but the cost of training (over 20 million parameters) and risk of overfitting persist. These limitations restrict the practical deployment of MMIF in real-time or edge-based medical devices like portable ultrasound or emergency CT units. Efforts to develop lightweight architectures, such as Mobile Net or efficient transformer hybrids, have been partially successful. For instance, Kalamkar & Geetha [89] propose transfer learning-based MMIF using pretrained models to reduce training time. However, performance degradation and sensitivity to modality shifts are still major issues. Hybrid DL models, such as PCNN + NSCT or GANs over NSST coefficients, further increase architectural complexity, which limits scalability and increases latency during clinical inference.
8.4. Ethical and Privacy Concerns in Multimodal Data Use
The use of medical images, especially in a multimodal and longitudinal context, raises significant privacy and ethical issues. Most MMIF datasets are derived from patients with chronic or terminal conditions, making de-identification difficult due to unique anatomical signatures (e.g., tumors or prostheses). Compliance with GDPR, HIPAA, and institutional IRB guidelines restricts dataset sharing. El-Shafai et al. [70] observed that fewer than 20% of fusion studies in recent years could access external datasets, limiting generalizability and leading to model bias. Federated learning, as explored in Goyal et al. [90], is a promising avenue to train models across silos without data exchange. However, challenges in harmonizing modalities, synchronizing update cycles, and addressing vulnerability to gradient attacks remain. Ethically, there's a lack of transparency in fusion models. Black-box CNNs may produce composite images that suppress or misrepresent subtle pathologies. There are also legal uncertainties in attributing diagnostic responsibility when AI-generated fusion images are used clinically. To address this, future MMIF research must include explainable AI (XAI) methods, probabilistic uncertainty maps, and formal ethical frameworks that define accountability for AI-assisted diagnostics [115]. While the field of MMIF continues to evolve rapidly, these four key challenges, data imbalance, registration inconsistency, computational overhead, and ethical constraints, remain persistent obstacles. Addressing these will require interdisciplinary collaboration between data scientists, radiologists, ethicists, and software engineers. The integration of robust registration models, edge-efficient architectures, federated training schemes, and ethically compliant data pipelines will be vital to unlocking MMIF’s full potential.
9. FUTURE RESEARCH DIRECTIONS IN MULTIMODAL MEDICAL IMAGE FUSION
9.1. Explainable AI (XAI) in MMIF
Explainable artificial intelligence is a technique for AI-powered diagnosis and analysis that ensures features such as ethics, transparency, and accountability in the traditional approach to AI. This will lead to outcome tracing and model improvements in health care. EXAI relies on feature extraction to make the model more explainable and interpretable. EXAI proposed a self-explanatory framework based on design principles for understanding and predicting the behavior of ML/DL models. Although deep models such as CNNs and Transformers outperform traditional algorithms, their “black-box” nature hinders clinical trust. Recent work integrates attention maps, Layer-wise Relevance Propagation (LRP), and Grad-CAMs into MMIF pipelines, allowing radiologists to verify the influence of fused features during diagnostics [115, 116].
9.2. Quantum Image Fusion
Quantum computation in medical image processing has improved edge detection, segmentation, watermarking, encryption, and classification. A quantum edge detection technique using superposition and fuzzy entropy can better detect strong and weak edges. Automatic hippocampal segmentation using a Quantum-Inspired Evolutionary Algorithm (QIEA) yielded good correlation between segmented and microscopic images. A hybrid technique using QPSO and fuzzy k-nearest neighbours enhances cervical cancer cell classification on the Herlev dataset for feature selection and classification. The hybrid method decreased features from 17 to 7, outperforming Naive Bayes and SVM with a 2% to 11% increase in accuracy. These inventions show how quantum computing might improve medical image analysis and diagnosis precision [117].
9.3. Federated and Privacy-preserving MMIF
Machine learning algorithms learn to solve problems autonomously based on the data provided to them, but they require extensive training to do so. Personal data in training datasets for AI systems, especially for health care applications, must be considered. Data from special categories (including health data) requires more safeguards than common data under the General Data Protection Regulation (GDPR). AI developers and users in health care are especially affected by this issue. With increasing data privacy regulations (e.g., GDPR, HIPAA), centralized MMIF training on patient data faces ethical and legal barriers. Federated learning frameworks (e.g., FL-MedFuseNet) enable MMIF model training across distributed hospital nodes without sharing raw data. This decentralized approach preserves privacy and supports continual learning from real-world clinical inputs [118].
9.4. Robotic Surgery and Smart Operating Rooms
The integration of MMIF into AI-assisted robotic surgeries is a fast-growing field. As the population ages, the prevalence of spinal degenerative illnesses such as lumbar disc herniation and spinal stenosis increases the need for accurate and safe spinal operations. The spine intricacy makes surgery difficult, making robotic help invaluable. Pedicle screw implantation is performed using surgical robots like TiRobot, Mazor, Da Vinci, ROSA, Excelsius GPS, and Orthbot, which enhance precision, reduce operative time, decrease the chances of hemorrhage, and minimize radiation exposure. These approaches use unimodal CT scans, which cannot detect nerves and intervertebral discs. The incorporation of a multimodal image fusion (CT/MR)- based software system for intraoperative navigation expands the horizons of robotic spinal surgery. Intuitive Surgical’s Da Vinci system already shows early adoption of MMIF-enhanced visual pipelines, although this is mostly experimental. Future research could focus on ultra-low-latency hardware-driven MMIF solutions embedded into surgical robots. NVIDIA’s Clara, a GPU-accelerated platform, is used to segment and diagnose cardiac images in real time. To improve processing speed and accuracy, the system takes advantage of powerful DL algorithms explicitly designed for cardiac imaging. Using rigorous data processing and a deep learning model, it can perform exact partitioning of cardiac components and identify positioning defects, resulting in the diagnosis of rapid heart arrhythmia [119, 120].
9.5. MMIF-based Watermarking for Tele-health Applications
Watermarking in medical image fusion involves embedding a hidden signal (such as patient ID, diagnosis detail, time stamps, or institutional information) into a fused image generated from multiple modalities. This ensures date integrity, ownership authentication, and secure communication in telemedicine or cloud environments. Embedding of watermark is done by techniques like Redundant Discrete Wavelet Transform (RDWT), Singular Value Decomposition (SVD), or NSCT. The integration of deep learning needs further exploration to create adaptive and intelligent watermarking methods [121].
CONCLUSION
This comprehensive review concludes that multimodal medical image fusion (MMIF) is a growing field with applications and modern techniques that allow the proper use of all available information, thus contributing to the improvement of clinical diagnostics. Firstly, the limitations of unimodal imaging were established, and the clinical need for combining MRI, CT, PET, and SPECT was explored. Combining strengths in each modality (by way of fusion) offers synergistic improvements to their diagnostic capability while addressing the limitations that each modality suffers from individually. The mentioned approaches, spatial and frequency-based fusion methods such as PCA, DWT, and NSCT, have proved useful in many early-stage applications owing to their simplicity and speed. They are sensitive to noise and often suffer from issues like edge blurring or spectral distortion. This led to the development of more advance strategies like sparse representation, which is more flexible, particularly under uncertain and noisy imaging conditions. Additionally, when applicable with multi-scale transforms, sparse coding improves edge structure and clarity that is necessary for tumor detection and neurodegenerative diseases. With the introduction of Deep Learning (DL) and hybrid fusion models in the field, a paradigm shift has occurred, drastically improving fusion accuracy and robustness. It is effectuated by various architectures such as CNNs, U-Nets, GANs, Swin Transformers, etc. Deep Learning has brought its weaknesses along, but it has also brought hybrid models that blend Deep Learning with usual domains to minimize and compensate for their respective weak points. Moreover, high-quality datasets (TCIA, ADNI, OASIS, AANLIB) and evaluation metrics (SSIM, MI, and entropy) are used as a standard benchmark for MMIF models. The results of experiments across two datasets in this review highlight the effectiveness of hybrid and deep learning-based approaches. Yet, there are still many outstanding issues. Finding its way into clinical practice is impeded by data scarcity, registration inconsistencies, the computational cost of DL models, and the ethical use of necessary patient data. The promising emerging topics of interest included federated learning, explainable AI, quantum fusion, and watermarking in telemedicine. The integration of MMIF into surgical robotics is another frontier that needs to be explored. Finally, the findings of this review offer a fundamental analytical lens with which healthcare professionals, AI researchers, and developers of MMIF systems can improve the clinical scalability, interpretability, and ethics of the systems that are generated. It can be concluded that the field is rapidly evolving and holds promise for improving clinical decision-making. MMIF had demonstrated enhanced diagnostic accuracy by combining information from multiple sources, improved treatment planning, and even direct benefits to patients through shorter and safer procedures. MMIF offers immense potential yet has limitations like registration errors, computational burdens, generation of artifacts, loss of specific information, and a lack of standardized evaluation metrics. Ultimately, ongoing interdisciplinary collaborations will significantly enhance precision and accuracy, improving patient outcomes in the years to come.
AUTHORS’ CONTRIBUTIONS
The authors confirm their contribution to the paper as follows: A.D.: Analysis and interpretation of results; S.B., B.G.: Writing - Reviewing and editing; N.G.: Writing - Original draft preparation. All authors reviewed the results and approved the final version of the manuscript.
LIST OF ABBREVIATIONS
| MMIF | = Multimodal Medical Image Fusion |
| MRI | = Magnetic Resonance Imaging |
| CT | = Computed Tomography |
| PET | = Positron Emission Tomography |
| SPECT | = Single Photon Emission Computed Tomography |
| fMRI | = Functional Magnetic Resonance Imaging |
| CBCT | = Cone Beam Computed Tomography |
| CAD | = Computer-Aided Diagnosis |
| TCIA | = The Cancer Imaging Archive |
| OASIS | = Open Access Series of Imaging Studies |
| ADNI | = Alzheimer’s Disease Neuroimaging Initiative |
| MIDAS | = Medical Image Data Archive System |
| AANLIB | = Annotated Alzheimer Neuroimaging Library |
| DDSM | = Digital Database for Screening Mammography |
| FLAIR | = Fluid-Attenuated Inversion Recovery |
| CSF | = Cerebrospinal Fluid |
| API | = Average Pixel Intensity |
| SD | = Standard Deviation |
| AG | = Average Gradient |
| EN | = Entropy |
| MI | = Mutual Information |
| FS | = Information Symmetry |
| CC | = Correlation Coefficient |
| SF | = Spatial Frequency |
| PSNR | = Peak Signal-to-Noise Ratio |
| MSE | = Mean Squared Error |
| SSIM | = Structural Similarity Index |
| VIF | = Visual Information Fidelity |
| SCD | = Spatial Correlation Difference |
| Qabf | = Edge-Based Similarity Measure |
| Nabf | = Noise-Based Similarity Measure |
| PCA | = Principal Component Analysis |
| ICA | = Independent Component Analysis |
| IHS | = Intensity-Hue-Saturation |
| DWT | = Discrete Wavelet Transform |
| NSCT | = Non-Subsampled Contourlet Transform |
| NSST | = Non-Subsampled Shearlet Transform |
| DFRWT | = Discrete Fractional Wavelet Transform |
| CSR | = Convolutional Sparse Representation |
| JSR | = Joint Sparse Representation |
| MCA | = Morphological Component Analysis |
| CS-MCA | = Convolutional Sparse Morphological Component Analysis |
| CNN | = Convolutional Neural Network |
| GAN | = Generative Adversarial Network |
| U-Net | = U-shaped Convolutional Neural Network |
| PCNN | = Pulse-Coupled Neural Network |
| ANFIS | = Adaptive Neuro-Fuzzy Inference System |
| IST | = Improved Structure Tensor |
| VMD | = Variational Mode Decomposition |
AVAILABILITY OF DATA AND MATERIALS
Authors may deposit their datasets openly to Zenodo Repository, in addition to their own or their institutional archives [114]. For more details please visit https://openneuroimagingjournal.com/
CONFLICT OF INTEREST
Dr. Ayush Dogra is the Editorial Advisory Board of The Open Neuroimaging Journal.
ACKNOWLEDGEMENTS
Declared none.


